| Identification |
|---|
| YMDB ID | YMDB00548 |
|---|
| Name | zymosterol |
|---|
| Species | Saccharomyces cerevisiae |
|---|
| Strain | Baker's yeast |
|---|
| Description | Zymosterol intermediate 2, also known as zymostrol or 5a-cholesta-8,24-dien-3b-ol, belongs to the class of organic compounds known as cholesterols and derivatives. Cholesterols and derivatives are compounds containing a 3-hydroxylated cholestane core. Zymosterol intermediate 2 is a very hydrophobic molecule, practically insoluble (in water), and relatively neutral. |
|---|
| Structure | |
|---|
| Synonyms | - 5-alpha-cholesta-8,24-dien-3-beta-ol
- 5alpha-Cholesta-8,24-dien-3beta-ol
- delta8,24-Cholestadien-3beta-ol
- Zymostrol
- 5a-Cholesta-8,24-dien-3b-ol
- 5Α-cholesta-8,24-dien-3β-ol
- delta8,24-Cholestadien-3b-ol
- Δ8,24-cholestadien-3β-ol
- Zymosterol intermediic acid 2
- Δ8,24-cholestadien-3b-ol
- (3b,5a)- Cholesta-8,24-dien-3-ol
- Cholest-8,24-dien-3b-ol
- Cholesta-8,24-dien-3-ol
- Zymosterol
|
|---|
| CAS number | 7488-99-5 |
|---|
| Weight | Average: 384.6377
Monoisotopic: 384.33921603 |
|---|
| InChI Key | CGSJXLIKVBJVRY-XTGBIJOFSA-N |
|---|
| InChI | InChI=1S/C27H44O/c1-18(2)7-6-8-19(3)23-11-12-24-22-10-9-20-17-21(28)13-15-26(20,4)25(22)14-16-27(23,24)5/h7,19-21,23-24,28H,6,8-17H2,1-5H3/t19-,20+,21+,23-,24+,26+,27-/m1/s1 |
|---|
| IUPAC Name | (2S,5S,7S,11R,14R,15R)-2,15-dimethyl-14-[(2R)-6-methylhept-5-en-2-yl]tetracyclo[8.7.0.0^{2,7}.0^{11,15}]heptadec-1(10)-en-5-ol |
|---|
| Traditional IUPAC Name | (2S,5S,7S,11R,14R,15R)-2,15-dimethyl-14-[(2R)-6-methylhept-5-en-2-yl]tetracyclo[8.7.0.0^{2,7}.0^{11,15}]heptadec-1(10)-en-5-ol |
|---|
| Chemical Formula | C27H44O |
|---|
| SMILES | [H][C@@]1(CC[C@@]2([H])C3=C(CC[C@]12C)[C@@]1(C)CC[C@H](O)C[C@]1([H])CC3)[C@H](C)CCC=C(C)C |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as cholesterols and derivatives. Cholesterols and derivatives are compounds containing a 3-hydroxylated cholestane core. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Steroids and steroid derivatives |
|---|
| Sub Class | Cholestane steroids |
|---|
| Direct Parent | Cholesterols and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Cholesterol-skeleton
- 3-beta-hydroxysteroid
- Hydroxysteroid
- 3-hydroxysteroid
- Cyclic alcohol
- Secondary alcohol
- Organic oxygen compound
- Hydrocarbon derivative
- Organooxygen compound
- Alcohol
- Aliphatic homopolycyclic compound
|
|---|
| Molecular Framework | Aliphatic homopolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Charge | 0 |
|---|
| Melting point | 187.5 °C |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Water Solubility | Not Available | PhysProp | | LogP | Not Available | PhysProp |
|
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations | - extracellular
- cell envelope
- lipid particle
- endoplasmic reticulum
- cytoplasm
|
|---|
| Organoleptic Properties | Not Available |
|---|
| SMPDB Pathways | | Cholesterol biosynthesis and metabolism CE(10:0) | PW002545 | 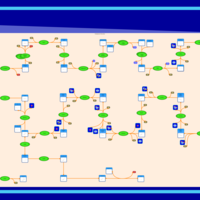 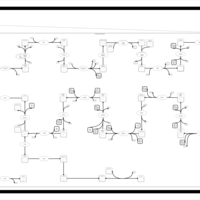 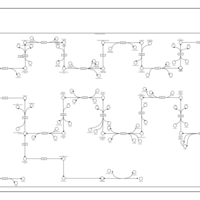 | | Cholesterol biosynthesis and metabolism CE(12:0) | PW002548 | 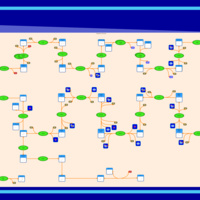   | | Cholesterol biosynthesis and metabolism CE(14:0) | PW002544 |   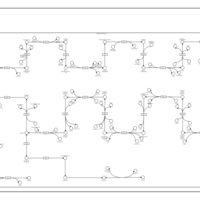 | | Cholesterol biosynthesis and metabolism CE(16:0) | PW002550 | 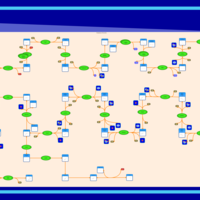 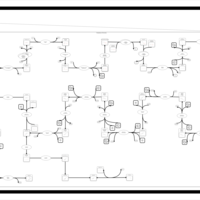 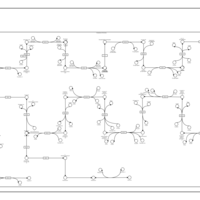 | | Cholesterol biosynthesis and metabolism CE(18:0) | PW002551 |    |
|
|---|
| KEGG Pathways | |
|---|
| SMPDB Reactions | |
|---|
| KEGG Reactions |
|
|---|
| Concentrations |
|---|
| Intracellular Concentrations | Not Available |
|---|
| Extracellular Concentrations | Not Available |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0cfr-1019000000-1b05483747ddc32500b0 | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-002f-3106900000-eda3cfead337ce02b2b5 | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014r-0019000000-010b245dedfb160716ba | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-014i-2149000000-c0828080ef844c1d5c69 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-02vi-3259000000-96993d821222323ab075 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001i-0009000000-0373115d11504acc8b81 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-001i-0009000000-cc742e0af833cc290a8f | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0gb9-1019000000-87e4aab6501a99f5baec | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001i-0009000000-4d7a5f3b63c0fb62f1d7 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-001i-0009000000-4d7a5f3b63c0fb62f1d7 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-001i-0019000000-384e7aa03192295e1589 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-0019000000-3fe44ab0178bb12e2a16 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-001i-4249000000-ee1fd80800f89af74a91 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0abc-8930000000-32bc5a989db5c704eabf | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer |
|
|---|
| References |
|---|
| References: | - UniProt Consortium (2011). "Ongoing and future developments at the Universal Protein Resource." Nucleic Acids Res 39:D214-D219.21051339
- Herrgard, M. J., Swainston, N., Dobson, P., Dunn, W. B., Arga, K. Y., Arvas, M., Bluthgen, N., Borger, S., Costenoble, R., Heinemann, M., Hucka, M., Le Novere, N., Li, P., Liebermeister, W., Mo, M. L., Oliveira, A. P., Petranovic, D., Pettifer, S., Simeonidis, E., Smallbone, K., Spasic, I., Weichart, D., Brent, R., Broomhead, D. S., Westerhoff, H. V., Kirdar, B., Penttila, M., Klipp, E., Palsson, B. O., Sauer, U., Oliver, S. G., Mendes, P., Nielsen, J., Kell, D. B. (2008). "A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology." Nat Biotechnol 26:1155-1160.18846089
- Wilcox, L. J., Balderes, D. A., Wharton, B., Tinkelenberg, A. H., Rao, G., Sturley, S. L. (2002). "Transcriptional profiling identifies two members of the ATP-binding cassette transporter superfamily required for sterol uptake in yeast." J Biol Chem 277:32466-32472.12077145
- Koffel, R., Tiwari, R., Falquet, L., Schneiter, R. (2005). "The Saccharomyces cerevisiae YLL012/YEH1, YLR020/YEH2, and TGL1 genes encode a novel family of membrane-anchored lipases that are required for steryl ester hydrolysis." Mol Cell Biol 25:1655-1668.15713625
- Zweytick, D., Leitner, E., Kohlwein, S. D., Yu, C., Rothblatt, J., Daum, G. (2000). "Contribution of Are1p and Are2p to steryl ester synthesis in the yeast Saccharomyces cerevisiae." Eur J Biochem 267:1075-1082.10672016
|
|---|
| Synthesis Reference: | Not Available |
|---|
| External Links: | |
|---|

