| Identification |
|---|
| YMDB ID | YMDB00330 |
|---|
| Name | L-Lysine |
|---|
| Species | Saccharomyces cerevisiae |
|---|
| Strain | Baker's yeast |
|---|
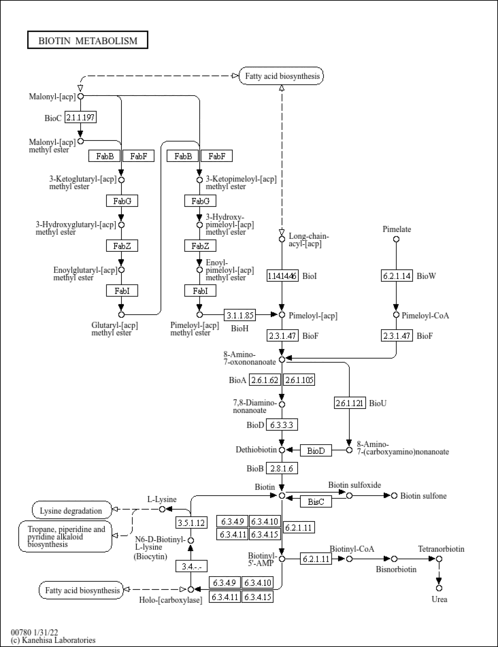
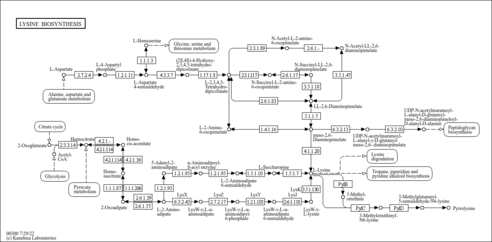
| Description | Lysine (abbreviated as Lys or K) is an alpha-amino acid. The L-isomer is one of the 22 proteinogenic amino acids, i.e., the building blocks of proteins. It is classified as a polar, basic amino acid, because of the positively charged nature of the amino-alkyl side chain. In yeast, lysine is synthesized from aspartic acid (aspartate). Specifically, L-aspartate is first converted to L-aspartyl-4-phosphate by aspartokinase (or Aspartate kinase). Subsequently, beta-aspartate semialdehyde dehydrogenase converts this into beta-aspartyl-4-semialdehyde (or beta-aspartate-4-semialdehyde) through an NADH-dependent reaction. After this step, Dihydrodipicolinate synthase adds a pyruvate group to the beta-aspartyl-4-semialdehyde, and two water molecules are removed. This causes cyclization and gives rise to 2,3-dihydrodipicolinate. This product is reduced to 2,3,4,5-tetrahydrodipicolinate by dihydrodipicolinate reductase. Tetrahydrodipicolinate N-acetyltransferase opens this ring and gives rise to N-succinyl-L-2-amino-6-oxoheptanedionate (or N-acyl-2-amino-6-oxopimelate). N-succinyl-L-2-amino-6-oxoheptanedionate is converted into N-succinyl-LL-2,6-diaminoheptanedionate (N-acyl-2,6-diaminopimelate). This reaction is catalyzed by the enzyme succinyl diaminopimelate aminotransferase. A glutaric acid molecule is used in this reaction and an oxoacid is produced as a byproduct. N-succinyl-LL-2,6-diaminoheptanedionate (N-acyl-2,6-diaminopimelate)is converted into LL-2,6-diaminoheptanedionate (L,L-2,6-diaminopimelate) by succinyl diaminopimelate desuccinylase (acyldiaminopimelate deacylase). A water molecule is consumed in this reaction and a succinate is produced a byproduct. LL-2,6-diaminoheptanedionate is converted by diaminopimelate epimerase into meso-2,6-diamino-heptanedionate (meso-2,6-diaminopimelate). Finally meso-2,6-diamino-heptanedionate is converted into L-lysine by diaminopimelate decarboxylase. Lysine catabolism is unusual in the way that the ε-amino group is transferred to alpha-ketoglutarate and into the general nitrogen pool. The reaction is a transamination in which the ε-amino group is transferred to the alpha-keto carbon of alpha-ketoglutarate forming the metabolite, saccharopine. Saccharopine is immediately hydrolyzed by the enzyme alpha-aminoadipic semialdehyde synthase in such a way that the amino nitrogen remains with the alpha-carbon of alpha-ketoglutarate, producing glutamate and alpha-aminoadipic semialdehyde. The ultimate end-product of lysine catabolism is acetoacetyl-CoA. |
|---|
| Structure | |
|---|
| Synonyms | - (+)-S-Lysine
- (S)-2,6-diamino-Hexanoate
- (S)-2,6-diamino-Hexanoic acid
- (S)-2,6-Diaminohexanoate
- (S)-2,6-Diaminohexanoic acid
- (S)-a,e-Diaminocaproate
- (S)-a,e-Diaminocaproic acid
- (S)-Lysine
- 2,6-Diaminohexanoate
- 2,6-Diaminohexanoic acid
- 6-amino-Aminutrin
- 6-amino-L-Norleucine
- a-Lysine
- alpha-Lysine
- Aminutrin
- h-Lys-oh
- L-(+)-Lysine
- L-2,6-Diainohexanoate
- L-2,6-Diainohexanoic acid
- L-2,6-Diaminocaproate
- L-2,6-Diaminocaproic acid
- L-Lys
- Lys
- Lysine
- Lysine acid
- (S)-alpha,epsilon-Diaminocaproic acid
- 6-Ammonio-L-norleucine
- K
- L-Lysin
- Lysina
- Lysinum
- (S)-a,epsilon-Diaminocaproate
- (S)-a,epsilon-Diaminocaproic acid
- (S)-alpha,epsilon-Diaminocaproate
- (S)-Α,epsilon-diaminocaproate
- (S)-Α,epsilon-diaminocaproic acid
- Acetate, lysine
- Enisyl
- Lysine hydrochloride
- L Lysine
- Lysine acetate
|
|---|
| CAS number | 56-87-1 |
|---|
| Weight | Average: 146.1876
Monoisotopic: 146.105527702 |
|---|
| InChI Key | KDXKERNSBIXSRK-YFKPBYRVSA-N |
|---|
| InChI | InChI=1S/C6H14N2O2/c7-4-2-1-3-5(8)6(9)10/h5H,1-4,7-8H2,(H,9,10)/t5-/m0/s1 |
|---|
| IUPAC Name | (2S)-2,6-diaminohexanoic acid |
|---|
| Traditional IUPAC Name | L-lysine |
|---|
| Chemical Formula | C6H14N2O2 |
|---|
| SMILES | [H]OC(=O)[C@@]([H])(N([H])[H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])N([H])[H] |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as l-alpha-amino acids. These are alpha amino acids which have the L-configuration of the alpha-carbon atom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | L-alpha-amino acids |
|---|
| Alternative Parents | |
|---|
| Substituents | - L-alpha-amino acid
- Medium-chain fatty acid
- Amino fatty acid
- Fatty acid
- Fatty acyl
- Amino acid
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Organic oxide
- Organopnictogen compound
- Organic oxygen compound
- Primary amine
- Organooxygen compound
- Organonitrogen compound
- Primary aliphatic amine
- Carbonyl group
- Organic nitrogen compound
- Amine
- Hydrocarbon derivative
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Charge | 0 |
|---|
| Melting point | 224.5 °C |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Water Solubility | 1000 mg/mL at 20 oC [YALKOWSKY,SH & DANNENFELSER,RM (1992)] | PhysProp | | LogP | -3.05 [HANSCH,C ET AL. (1995)] | PhysProp |
|
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations | - extracellular
- mitochondrion
- vacuole
- cytoplasm
|
|---|
| Organoleptic Properties | |
|---|
| SMPDB Pathways | |
|---|
| KEGG Pathways | |
|---|
| SMPDB Reactions | |
|---|
| KEGG Reactions | |
|---|
| Concentrations |
|---|
| Intracellular Concentrations | | Intracellular Concentration | Substrate | Growth Conditions | Strain | Citation |
|---|
| 702 ± 14 µM | YPD media | aerobic | Baker's yeast | PMID: 7654310 | | 561 ± 11 µM | YPG media | aerobic | Baker's yeast | PMID: 7654310 | | 351 ± 7 µM | SD media | aerobic | Baker's yeast | PMID: 7654310 | | 421 ± 8 µM | SG media | aerobic | Baker's yeast | PMID: 7654310 | | 351 ± 7 µM | M (molasses) | aerobic | Baker's yeast | PMID: 7654310 | | 842 ± 17 µM | MA (molasses) | aerobic | Baker's yeast | PMID: 7654310 | | 281 ± 6 µM | MB (molasses) | aerobic | Baker's yeast | PMID: 7654310 | | 772 ± 15 µM | MAB (molasses) | aerobic | Baker's yeast | PMID: 7654310 | | 3620 ± 181 µM | YEB media with 0.5 mM glucose | aerobic | Baker's yeast | Experimentally Determined
Not Available | | 1860 ± 683 µM | Synthetic medium with 20 g/L glucose | aerobic | Baker's yeast | PMID: 12584756 | | Conversion Details Here |
|
|---|
| Extracellular Concentrations | Not Available |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (3 TMS) | splash10-00di-3910000000-98c565675de67aa87900 | JSpectraViewer | MoNA | | GC-MS | GC-MS Spectrum - GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (Non-derivatized) | splash10-0ab9-1910000000-87ef8534f592041f50f2 | JSpectraViewer | MoNA | | GC-MS | GC-MS Spectrum - GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (4 TMS) | splash10-0a4i-1921000000-84f7815b0f650fa17444 | JSpectraViewer | MoNA | | GC-MS | GC-MS Spectrum - GC-MS (3 TMS) | splash10-001i-9600000000-823408dba509cb204acf | JSpectraViewer | MoNA | | GC-MS | GC-MS Spectrum - GC-MS (3 TMS) | splash10-00di-3910000000-4f5578af5e7d8b6c49f7 | JSpectraViewer | MoNA | | GC-MS | GC-MS Spectrum - GC-MS (4 TMS) | splash10-0adi-1921000000-4e56d95e623e792f9e6b | JSpectraViewer | MoNA | | GC-MS | GC-MS Spectrum - EI-B (Non-derivatized) | splash10-00di-0921000000-eeb49e57bc1a75193058 | JSpectraViewer | MoNA | | GC-MS | GC-MS Spectrum - EI-B (Non-derivatized) | splash10-0ab9-0921000000-ebb902be0f3754225b2f | JSpectraViewer | MoNA | | GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-00di-3910000000-98c565675de67aa87900 | JSpectraViewer | MoNA | | GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-0ab9-1910000000-87ef8534f592041f50f2 | JSpectraViewer | MoNA | | GC-MS | GC-MS Spectrum - GC-EI-QQ (Non-derivatized) | splash10-0fdk-3923000000-15b84c2649c1b0455de1 | JSpectraViewer | MoNA | | GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-0a4i-1921000000-84f7815b0f650fa17444 | JSpectraViewer | MoNA | | GC-MS | GC-MS Spectrum - GC-MS (Non-derivatized) | splash10-001i-9600000000-823408dba509cb204acf | JSpectraViewer | MoNA | | GC-MS | GC-MS Spectrum - GC-MS (Non-derivatized) | splash10-00di-3910000000-4f5578af5e7d8b6c49f7 | JSpectraViewer | MoNA | | GC-MS | GC-MS Spectrum - GC-MS (Non-derivatized) | splash10-0adi-1921000000-4e56d95e623e792f9e6b | JSpectraViewer | MoNA | | GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-0abi-1900000000-9ad174122e4d6e003eb8 | JSpectraViewer | MoNA | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0089-9100000000-974cc55c9130ed5213eb | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-0ue9-9700000000-57b24ae819b6ec26bfd2 | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_2) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_3) - 70eV, Positive | Not Available | JSpectraViewer | | LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-003r-8900000000-470a0beb4f338ed89bca | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-001i-9000000000-74e9193d9d33c2509bfa | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-0a59-9000000000-822c4e78250fffa56e39 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0002-0900000000-04f9a62a77fb5a37ca22 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-001i-9000000000-035035ecfa084671479b | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-001i-0900000000-5bb15839f86f4fca0d0b | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-001i-0900000000-6c8ef03aa83eb1cab35b | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-01ot-0910000000-c182a7dcdbc260666978 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0a4j-0900000000-6c5f378cef2f14204e15 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-001i-0900000000-41e1a6499097748934b0 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-03di-0390000000-7270a0b85b9e3f9f6373 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Negative | splash10-0002-0900000000-a997e809874357908880 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Negative | splash10-0002-2900000000-f64110414f82c93e1fe8 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Negative | splash10-014m-9200000000-33a38e6370811c5ddc82 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Negative | splash10-0006-9000000000-3a8b0b6e62f5c66d3720 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 50V, Negative | splash10-0006-9000000000-d5167570d11d77fd541e | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Positive | splash10-0002-0900000000-a46231bd529176101129 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Positive | splash10-001i-9200000000-f8b9f01b2a9886c51df9 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Positive | splash10-001i-9000000000-ace0361476d939043e98 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Positive | splash10-001i-9000000000-5b06b6e9dcf9faf8e89c | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 50V, Positive | splash10-053r-9000000000-2894ef6d7f72ea71688e | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - CE-ESI-TOF (CE-system connected to 6210 Time-of-Flight MS, Agilent) , Positive | splash10-0002-0900000000-290902f43cf851e8ef5e | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Positive | splash10-001i-9300000000-f81193f6b50235ec8147 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Positive | splash10-001i-9300000000-8931d2193adab166d5e4 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Negative | splash10-0002-0900000000-b4825cc64fcb830c6967 | JSpectraViewer | MoNA | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 2D NMR | [1H,1H] 2D NMR Spectrum | Not Available | JSpectraViewer | | 2D NMR | [1H,13C] 2D NMR Spectrum | Not Available | JSpectraViewer |
|
|---|
| References |
|---|
| References: | - Martinez-Force, E., Benitez, T. (1995). "Effects of varying media, temperature, and growth rates on the intracellular concentrations of yeast amino acids." Biotechnol Prog 11:386-392.7654310
- UniProt Consortium (2011). "Ongoing and future developments at the Universal Protein Resource." Nucleic Acids Res 39:D214-D219.21051339
- Scheer, M., Grote, A., Chang, A., Schomburg, I., Munaretto, C., Rother, M., Sohngen, C., Stelzer, M., Thiele, J., Schomburg, D. (2011). "BRENDA, the enzyme information system in 2011." Nucleic Acids Res 39:D670-D676.21062828
- Herrgard, M. J., Swainston, N., Dobson, P., Dunn, W. B., Arga, K. Y., Arvas, M., Bluthgen, N., Borger, S., Costenoble, R., Heinemann, M., Hucka, M., Le Novere, N., Li, P., Liebermeister, W., Mo, M. L., Oliveira, A. P., Petranovic, D., Pettifer, S., Simeonidis, E., Smallbone, K., Spasic, I., Weichart, D., Brent, R., Broomhead, D. S., Westerhoff, H. V., Kirdar, B., Penttila, M., Klipp, E., Palsson, B. O., Sauer, U., Oliver, S. G., Mendes, P., Nielsen, J., Kell, D. B. (2008). "A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology." Nat Biotechnol 26:1155-1160.18846089
- Arevalo-Rodriguez, M., Calderon, I. L., Holmberg, S. (1999). "Mutations that cause threonine sensitivity identify catalytic and regulatory regions of the aspartate kinase of Saccharomyces cerevisiae." Yeast 15:1331-1345.10509015
- Hans, M. A., Heinzle, E., Wittmann, C. (2003). "Free intracellular amino acid pools during autonomous oscillations in Saccharomyces cerevisiae." Biotechnol Bioeng 82:143-151.12584756
- Castrillo, J. I., Zeef, L. A., Hoyle, D. C., Zhang, N., Hayes, A., Gardner, D. C., Cornell, M. J., Petty, J., Hakes, L., Wardleworth, L., Rash, B., Brown, M., Dunn, W. B., Broadhurst, D., O'Donoghue, K., Hester, S. S., Dunkley, T. P., Hart, S. R., Swainston, N., Li, P., Gaskell, S. J., Paton, N. W., Lilley, K. S., Kell, D. B., Oliver, S. G. (2007). "Growth control of the eukaryote cell: a systems biology study in yeast." J Biol 6:4.17439666
|
|---|
| Synthesis Reference: | Rothstein, Morton. DL-Lysine-6-C14 and DL-a-aminoadipic acid-6-C14. Biochemical Preparations (1961), 8 85-8. |
|---|
| External Links: | |
|---|