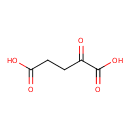
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (2 TMS) | splash10-000b-3910000000-4097b7eb52c2910a4277 | JSpectraViewer | MoNA | | GC-MS | GC-MS Spectrum - GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (Non-derivatized) | splash10-0002-1910000000-3277227b9c4da4baa1f7 | JSpectraViewer | MoNA | | GC-MS | GC-MS Spectrum - GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (2 TMS; 1 MEOX) | splash10-00di-9300000000-974bce546caa4fbe6b0f | JSpectraViewer | MoNA | | GC-MS | GC-MS Spectrum - GC-MS (1 MEOX; 2 TMS) | splash10-000i-9710000000-d0aea11e947b042571f9 | JSpectraViewer | MoNA | | GC-MS | GC-MS Spectrum - GC-MS (1 MEOX; 2 TMS) | splash10-052s-4920000000-bf7ed92c68fd4c8dbb6b | JSpectraViewer | MoNA | | GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-000b-3910000000-4097b7eb52c2910a4277 | JSpectraViewer | MoNA | | GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-0002-1910000000-3277227b9c4da4baa1f7 | JSpectraViewer | MoNA | | GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-00di-9300000000-974bce546caa4fbe6b0f | JSpectraViewer | MoNA | | GC-MS | GC-MS Spectrum - GC-MS (Non-derivatized) | splash10-000i-9710000000-d0aea11e947b042571f9 | JSpectraViewer | MoNA | | GC-MS | GC-MS Spectrum - GC-MS (Non-derivatized) | splash10-052s-4920000000-bf7ed92c68fd4c8dbb6b | JSpectraViewer | MoNA | | GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-000b-2900000000-e514f7afea2490bbe9ed | JSpectraViewer | MoNA | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0kfx-9200000000-0ee77a39e9862659f8a7 | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-00di-9730000000-5e17bd8cfd944963c79f | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_2) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_3) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_1) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_2) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_3) - 70eV, Positive | Not Available | JSpectraViewer | | LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Negative (Annotated) | splash10-0zfr-6900000000-7fc343a243ce5d2a4f39 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Negative (Annotated) | splash10-0a4i-9200000000-1a7af9946604baa8b3d2 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Negative (Annotated) | splash10-0a4l-9800000000-30495b2afdd3526854e7 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-0002-0925200000-af33449c9638276466c2 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-0a4i-9000000000-6e43f767847790112b65 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-03di-0900000000-4cacc41705b7b385e91e | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-0udi-0900000000-1367eeeb6ad64c67a0aa | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-000t-0915051100-91cda631ee54800322b8 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-0a4i-9000000000-4f842c3673925d29c402 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-0udi-0900000000-5e751351656f6eb62de3 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-001i-0910000000-7729faa24733fbad77b7 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Negative | splash10-0002-0900000000-c3c62f83bff99f44f7ab | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Negative | splash10-0zfr-6900000000-b6d8379db3e3f1480b14 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Negative | splash10-0a4i-9300000000-4fde39bb93cde5a27f23 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Negative | splash10-00i0-9000000000-4343832f3a55842423a9 | JSpectraViewer | MoNA | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-004i-2900000000-5aa3d6d620db1d733a65 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0m30-7900000000-fc77f69ad43f915bf23e | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a6r-9000000000-c597f1f5e416037aa82d | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-004i-2900000000-5aa3d6d620db1d733a65 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0m30-7900000000-fc77f69ad43f915bf23e | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a6r-9000000000-c597f1f5e416037aa82d | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-1900000000-f2b463076a150a514f0c | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0f9t-8900000000-b2d3fa8d7ab01a220d9a | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-053r-9100000000-554bfbd6839bc376cba0 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-1900000000-f2b463076a150a514f0c | JSpectraViewer | | MS | Mass Spectrum (Electron Ionization) | splash10-0fba-9200000000-4e367c66e94c51eb1a1b | JSpectraViewer | MoNA | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 2D NMR | [1H,13C] 2D NMR Spectrum | Not Available | JSpectraViewer |
|
|---|
| References: | - UniProt Consortium (2011). "Ongoing and future developments at the Universal Protein Resource." Nucleic Acids Res 39:D214-D219.21051339
- Scheer, M., Grote, A., Chang, A., Schomburg, I., Munaretto, C., Rother, M., Sohngen, C., Stelzer, M., Thiele, J., Schomburg, D. (2011). "BRENDA, the enzyme information system in 2011." Nucleic Acids Res 39:D670-D676.21062828
- Herrgard, M. J., Swainston, N., Dobson, P., Dunn, W. B., Arga, K. Y., Arvas, M., Bluthgen, N., Borger, S., Costenoble, R., Heinemann, M., Hucka, M., Le Novere, N., Li, P., Liebermeister, W., Mo, M. L., Oliveira, A. P., Petranovic, D., Pettifer, S., Simeonidis, E., Smallbone, K., Spasic, I., Weichart, D., Brent, R., Broomhead, D. S., Westerhoff, H. V., Kirdar, B., Penttila, M., Klipp, E., Palsson, B. O., Sauer, U., Oliver, S. G., Mendes, P., Nielsen, J., Kell, D. B. (2008). "A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology." Nat Biotechnol 26:1155-1160.18846089
- Roon, R. J., Even, H. L., Larimore, F. (1974). "Glutamate synthase: properties of the reduced nicotinamide adenine dinucleotide-dependent enzyme from Saccharomyces cerevisiae." J Bacteriol 118:89-95.4362465
- Takada, Y., Noguchi, T. (1985). "Characteristics of alanine: glyoxylate aminotransferase from Saccharomyces cerevisiae, a regulatory enzyme in the glyoxylate pathway of glycine and serine biosynthesis from tricarboxylic acid-cycle intermediates." Biochem J 231:157-163.3933486
- Hans, M. A., Heinzle, E., Wittmann, C. (2003). "Free intracellular amino acid pools during autonomous oscillations in Saccharomyces cerevisiae." Biotechnol Bioeng 82:143-151.12584756
- Moreira dos Santos, M., Thygesen, G., Kotter, P., Olsson, L., Nielsen, J. (2003). "Aerobic physiology of redox-engineered Saccharomyces cerevisiae strains modified in the ammonium assimilation for increased NADPH availability." FEMS Yeast Res 4:59-68.14554197
- Hogan, D. A., Auchtung, T. A., Hausinger, R. P. (1999). "Cloning and characterization of a sulfonate/alpha-ketoglutarate dioxygenase from Saccharomyces cerevisiae." J Bacteriol 181:5876-5879.10482536
- Bhattacharjee, J. K. (1985). "alpha-Aminoadipate pathway for the biosynthesis of lysine in lower eukaryotes." Crit Rev Microbiol 12:131-151.3928261
- Cupp, J. R., McAlister-Henn, L. (1992). "Cloning and characterization of the gene encoding the IDH1 subunit of NAD(+)-dependent isocitrate dehydrogenase from Saccharomyces cerevisiae." J Biol Chem 267:16417-16423.1644826
- Verleur, N., Elgersma, Y., Van Roermund, C. W., Tabak, H. F., Wanders, R. J. (1997). "Cytosolic aspartate aminotransferase encoded by the AAT2 gene is targeted to the peroxisomes in oleate-grown Saccharomyces cerevisiae." Eur J Biochem 247:972-980.9288922
- Dickinson, J. R., Harrison, S. J., Hewlins, M. J. (1998). "An investigation of the metabolism of valine to isobutyl alcohol in Saccharomyces cerevisiae." J Biol Chem 273:25751-25756.9748245
- Belhumeur, P., Fortin, N., Clark, M. W. (1994). "A gene from Saccharomyces cerevisiae which codes for a protein with significant homology to the bacterial 3-phosphoserine aminotransferase." Yeast 10:385-389.8017107
- Minard, K. I., Jennings, G. T., Loftus, T. M., Xuan, D., McAlister-Henn, L. (1998). "Sources of NADPH and expression of mammalian NADP+-specific isocitrate dehydrogenases in Saccharomyces cerevisiae." J Biol Chem 273:31486-31493.9813062
- Przybyla-Zawislak, B., Gadde, D. M., Ducharme, K., McCammon, M. T. (1999). "Genetic and biochemical interactions involving tricarboxylic acid cycle (TCA) function using a collection of mutants defective in all TCA cycle genes." Genetics 152:153-166.10224250
- Valenzuela, L., Ballario, P., Aranda, C., Filetici, P., Gonzalez, A. (1998). "Regulation of expression of GLT1, the gene encoding glutamate synthase in Saccharomyces cerevisiae." J Bacteriol 180:3533-3540.9657994
- Iraqui, I., Vissers, S., Cartiaux, M., Urrestarazu, A. (1998). "Characterisation of Saccharomyces cerevisiae ARO8 and ARO9 genes encoding aromatic aminotransferases I and II reveals a new aminotransferase subfamily." Mol Gen Genet 257:238-248.9491083
- Kispal, G., Steiner, H., Court, D. A., Rolinski, B., Lill, R. (1996). "Mitochondrial and cytosolic branched-chain amino acid transaminases from yeast, homologs of the myc oncogene-regulated Eca39 protein." J Biol Chem 271:24458-24464.8798704
- Boer, V. M., Crutchfield, C. A., Bradley, P. H., Botstein, D., Rabinowitz, J. D. (2010). "Growth-limiting intracellular metabolites in yeast growing under diverse nutrient limitations." Mol Biol Cell 21:198-211.19889834
- Gancedo, J. M., Gancedo, C. (1973). "Concentrations of intermediary metabolites in yeast." Biochimie 55:205-211.4578278
- Lagunas, R., Gancedo, C. (1983). "Role of phosphate in the regulation of the Pasteur effect in Saccharomyces cerevisiae." Eur J Biochem 137:479-483.6229402
- Castrillo, J. I., Zeef, L. A., Hoyle, D. C., Zhang, N., Hayes, A., Gardner, D. C., Cornell, M. J., Petty, J., Hakes, L., Wardleworth, L., Rash, B., Brown, M., Dunn, W. B., Broadhurst, D., O'Donoghue, K., Hester, S. S., Dunkley, T. P., Hart, S. R., Swainston, N., Li, P., Gaskell, S. J., Paton, N. W., Lilley, K. S., Kell, D. B., Oliver, S. G. (2007). "Growth control of the eukaryote cell: a systems biology study in yeast." J Biol 6:4.17439666
|
|---|