D-Erythrose 4-phosphate (YMDB00223)
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| YMDB ID | YMDB00223 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name | D-Erythrose 4-phosphate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Species | Saccharomyces cerevisiae | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Strain | Baker's yeast | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | D-Erythrose 4-phosphate, also known as D-erythrose-4-p or 4-O-phosphono-D-erythrose, belongs to the class of organic compounds known as monosaccharide phosphates. These are monosaccharides comprising a phosphated group linked to the carbohydrate unit. D-Erythrose 4-phosphate is an extremely weak basic (essentially neutral) compound (based on its pKa). D-Erythrose 4-phosphate exists in all living species, ranging from bacteria to humans. xylulose 5-phosphate and D-erythrose 4-phosphate can be biosynthesized from β-D-fructose 6-phosphate and D-glyceraldehyde 3-phosphate; which is catalyzed by the enzyme transketolase. In yeast, D-erythrose 4-phosphate is involved in the metabolic pathway called the riboneogenesis pathway. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number | 585-18-2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Weight | Average: 200.0838 Monoisotopic: 200.008589154 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key | NGHMDNPXVRFFGS-IUYQGCFVSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI | InChI=1S/C4H9O7P/c5-1-3(6)4(7)2-11-12(8,9)10/h1,3-4,6-7H,2H2,(H2,8,9,10)/t3-,4+/m0/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name | [(2R,3R)-2,3-dihydroxy-4-oxobutoxy]phosphonic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name | 4-O-phosphono-D-erythrose | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula | C4H9O7P | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES | [H]O[C@@]([H])(C([H])=O)[C@]([H])(O[H])C([H])([H])OP(=O)(O[H])O[H] | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as monosaccharide phosphates. These are monosaccharides comprising a phosphated group linked to the carbohydrate unit. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organic oxygen compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Organooxygen compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Carbohydrates and carbohydrate conjugates | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Monosaccharide phosphates | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic acyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge | 0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Organoleptic Properties | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMPDB Pathways |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
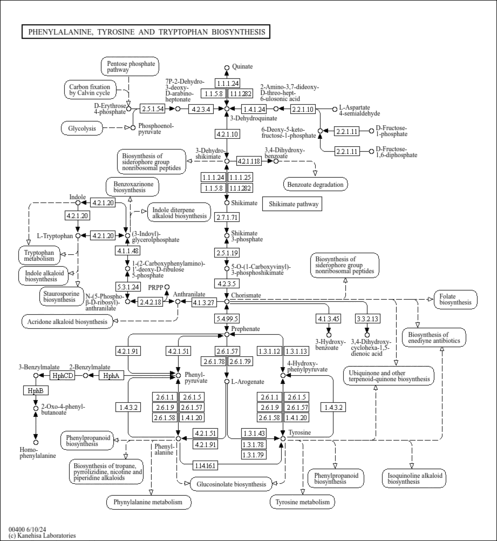
| KEGG Pathways |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMPDB Reactions |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG Reactions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Concentrations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Intracellular Concentrations | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Extracellular Concentrations | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in catalytic activity
- Specific function:
- Stereospecific condensation of phosphoenolpyruvate (PEP) and D-erythrose-4-phosphate (E4P) giving rise to 3-deoxy-D- arabino-heptulosonate-7-phosphate (DAHP)
- Gene Name:
- ARO4
- Uniprot ID:
- P32449
- Molecular weight:
- 39748.80078
Reactions
| Phosphoenolpyruvate + D-erythrose 4-phosphate + H(2)O → 3-deoxy-D-arabino-hept-2-ulosonate 7-phosphate + phosphate. |
- General function:
- Involved in catalytic activity
- Specific function:
- Stereospecific condensation of phosphoenolpyruvate (PEP) and D-erythrose-4-phosphate (E4P) giving rise to 3-deoxy-D- arabino-heptulosonate-7-phosphate (DAHP)
- Gene Name:
- ARO3
- Uniprot ID:
- P14843
- Molecular weight:
- 41069.5
Reactions
| Phosphoenolpyruvate + D-erythrose 4-phosphate + H(2)O → 3-deoxy-D-arabino-hept-2-ulosonate 7-phosphate + phosphate. |
- General function:
- Involved in catalytic activity
- Specific function:
- Sedoheptulose 7-phosphate + D-glyceraldehyde 3-phosphate = D-ribose 5-phosphate + D-xylulose 5-phosphate
- Gene Name:
- TKL2
- Uniprot ID:
- P33315
- Molecular weight:
- 75028.79688
Reactions
| Sedoheptulose 7-phosphate + D-glyceraldehyde 3-phosphate → D-ribose 5-phosphate + D-xylulose 5-phosphate. |
- General function:
- Involved in catalytic activity
- Specific function:
- Transaldolase is important for the balance of metabolites in the pentose-phosphate pathway
- Gene Name:
- NQM1
- Uniprot ID:
- P53228
- Molecular weight:
- 37253.30078
Reactions
| Sedoheptulose 7-phosphate + D-glyceraldehyde 3-phosphate → D-erythrose 4-phosphate + D-fructose 6-phosphate. |
- General function:
- Involved in catalytic activity
- Specific function:
- Sedoheptulose 7-phosphate + D-glyceraldehyde 3-phosphate = D-ribose 5-phosphate + D-xylulose 5-phosphate
- Gene Name:
- TKL1
- Uniprot ID:
- P23254
- Molecular weight:
- 73805.0
Reactions
| Sedoheptulose 7-phosphate + D-glyceraldehyde 3-phosphate → D-ribose 5-phosphate + D-xylulose 5-phosphate. |
- General function:
- Involved in catalytic activity
- Specific function:
- Catalyzes the aldol condensation of dihydroxyacetone phosphate (DHAP or glycerone-phosphate) with glyceraldehyde 3- phosphate (G3P) to form fructose 1,6-bisphosphate (FBP) in gluconeogenesis and the reverse reaction in glycolysis
- Gene Name:
- FBA1
- Uniprot ID:
- P14540
- Molecular weight:
- 39620.5
Reactions
| D-fructose 1,6-bisphosphate → glycerone phosphate + D-glyceraldehyde 3-phosphate. |
- General function:
- Involved in oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor
- Specific function:
- D-glyceraldehyde 3-phosphate + phosphate + NAD(+) = 3-phospho-D-glyceroyl phosphate + NADH
- Gene Name:
- TDH1
- Uniprot ID:
- P00360
- Molecular weight:
- 35749.60156
Reactions
| D-glyceraldehyde 3-phosphate + phosphate + NAD(+) → 3-phospho-D-glyceroyl phosphate + NADH. |
- General function:
- Involved in catalytic activity
- Specific function:
- Transaldolase is important for the balance of metabolites in the pentose-phosphate pathway
- Gene Name:
- TAL1
- Uniprot ID:
- P15019
- Molecular weight:
- 37036.0
Reactions
| Sedoheptulose 7-phosphate + D-glyceraldehyde 3-phosphate → D-erythrose 4-phosphate + D-fructose 6-phosphate. |