Homoaconitase, mitochondrial (P49367)
| Identification | |||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Name | Homoaconitase, mitochondrial | ||||||||||||||||||||||||||||||
| Synonyms |
| ||||||||||||||||||||||||||||||
| Gene Name | LYS4 | ||||||||||||||||||||||||||||||
| Enzyme Class | |||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||
| General Function | Involved in metabolic process | ||||||||||||||||||||||||||||||
| Specific Function | Responsible for the dehydration of cis-homoaconitate to homoisocitric acid | ||||||||||||||||||||||||||||||
| Cellular Location | Mitochondrion | ||||||||||||||||||||||||||||||
| SMPDB Pathways |
| ||||||||||||||||||||||||||||||
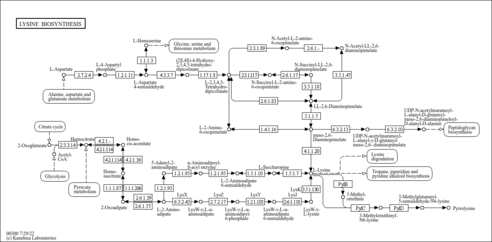
| KEGG Pathways |
| ||||||||||||||||||||||||||||||
| SMPDB Reactions |
| ||||||||||||||||||||||||||||||
| KEGG Reactions |
| ||||||||||||||||||||||||||||||
| Metabolites |
| ||||||||||||||||||||||||||||||
| GO Classification |
| ||||||||||||||||||||||||||||||
| Gene Properties | |||||||||||||||||||||||||||||||
| Chromosome Location | chromosome 4 | ||||||||||||||||||||||||||||||
| Locus | YDR234W | ||||||||||||||||||||||||||||||
| Gene Sequence | >2082 bp ATGCTACGATCAACCACATTTACTCGTTCGTTCCACAGTTCTAGGGCCTGGTTGAAAGGT CAGAACCTAACTGAAAAAATTGTTCAGTCGTATGCGGTCAACCTTCCCGAGGGTAAAGTT GTGCATTCTGGTGACTATGTATCGATCAAGCCGGCACACTGTATGTCCCACGATAATTCG TGGCCTGTAGCTTTGAAATTCATGGGGCTTGGCGCTACCAAGATCAAGAATCCTTCACAG ATTGTGACCACTCTGGACCACGATATTCAGAACAAATCAGAGAAAAATTTGACCAAGTAC AAGAACATCGAAAATTTTGCTAAGAAACACCATATAGACCACTACCCTGCCGGTAGAGGT ATTGGTCATCAAATTATGATTGAGGAGGGCTATGCTTTCCCCTTGAACATGACTGTCGCA TCTGACTCGCATTCAAACACCTACGGTGGTCTGGGGTCGCTGGGCACTCCAATAGTGAGA ACAGACGCTGCAGCCATATGGGCCACGGGACAGACGTGGTGGCAGATCCCACCAGTGGCT CAGGTTGAGTTGAAAGGTCAATTGCCTCAGGGTGTTTCCGGAAAAGATATCATTGTCGCA TTATGTGGGCTTTTCAACAATGATCAAGTTCTAAATCACGCCATTGAATTCACGGGTGAC TCTTTGAATGCATTGCCTATCGATCACAGACTCACTATTGCTAACATGACCACCGAGTGG GGGGCTCTTTCTGGTTTGTTCCCCGTGGACAAAACTTTGATCGACTGGTATAAAAACCGT TTGCAAAAGCTGGGCACCAATAATCATCCAAGGATTAATCCAAAGACTATCCGCGCACTA GAAGAAAAGGCGAAGATTCCGAAAGCAGACAAGGATGCACATTATGCCAAGAAACTGATC ATCGATCTAGCCACGCTAACTCACTACGTCTCAGGTCCAAATAGTGTTAAGGTCTCCAAC ACCGTGCAAGATCTATCTCAACAAGACATCAAGATAAATAAAGCTTATCTAGTGTCATGT ACAAACTCCCGTCTATCTGATTTGCAATCTGCAGCGGATGTGGTTTGTCCTACTGGAGAC TTAAACAAAGTCAACAAGGTGGCTCCAGGTGTGGAGTTCTATGTCGCTGCTGCCTCTTCA GAAATTGAGGCTGATGCCCGTAAATCAGGCGCTTGGGAAAAGCTGCTAAAGGCTGGCTGT ATCCCACTGCCTTCTGGTTGTGGTCCATGCATCGGTCTAGGTGCGGGATTACTGGAACCA GGTGAAGTTGGTATCAGTGCCACAAACAGAAACTTCAAAGGTAGAATGGGTTCCAAGGAT GCATTGGCTTACTTAGCTTCCCCTGCTGTAGTCGCCGCTTCTGCCGTGCTGGGTAAGATT AGTTCTCCTGCTGAGGTATTGTCCACAAGCGAAATTCCATTCAGCGGCGTTAAGACTGAG ATAATTGAGAATCCCGTGGTTGAAGAGGAAGTTAACGCTCAAACAGAGGCTCCAAAACAA TCCGTTGAGATATTAGAAGGTTTCCCAAGAGAGTTTTCTGGTGAATTAGTTTTATGTGAT GCCGATAACATCAATACCGATGGTATATATCCTGGTAAGTACACTTATCAGGATGATGTG CCTAAAGAAAAGATGGCGCAAGTTTGTATGGAAAATTATGATGCCGAGTTCAGAACCAAG GTTCATCCAGGTGATATAGTGGTCAGTGGGTTCAATTTCGGTACCGGTTCCTCCAGGGAA CGAGCGGCCACCGCCTTATTGGCTAAAGGTATCAACTTAGTTGTTTCAGGATCTTTTGGT AATATTTTTTCAAGAAACTCCATTAACAATGCTCTTCTGACCTTGGAAATCCCAGCATTA ATCAAAAAATTACGTGAGAAATATCAAGGTGCTCCAAAAGAACTTACAAGAAGAACTGGT TGGTTTTTGAAATGGGATGTAGCTGATGCTAAAGTGGTCGTTACCGAAGGTTCTTTGGAC GGCCCTGTGATCTTGGAGCAAAAAGTGGGTGAGCTAGGTAAGAACCTACAAGAAATTATT GTAAAAGGAGGCTTGGAAGGTTGGGTCAAATCCCAACTATAA | ||||||||||||||||||||||||||||||
| Protein Properties | |||||||||||||||||||||||||||||||
| Pfam Domain Function | |||||||||||||||||||||||||||||||
| Protein Residues | 693 | ||||||||||||||||||||||||||||||
| Protein Molecular Weight | 75150.10156 | ||||||||||||||||||||||||||||||
| Protein Theoretical pI | 7.41 | ||||||||||||||||||||||||||||||
| Signalling Regions |
| ||||||||||||||||||||||||||||||
| Transmembrane Regions |
| ||||||||||||||||||||||||||||||
| Protein Sequence | >Homoaconitase, mitochondrial MLRSTTFTRSFHSSRAWLKGQNLTEKIVQSYAVNLPEGKVVHSGDYVSIKPAHCMSHDNS WPVALKFMGLGATKIKNPSQIVTTLDHDIQNKSEKNLTKYKNIENFAKKHHIDHYPAGRG IGHQIMIEEGYAFPLNMTVASDSHSNTYGGLGSLGTPIVRTDAAAIWATGQTWWQIPPVA QVELKGQLPQGVSGKDIIVALCGLFNNDQVLNHAIEFTGDSLNALPIDHRLTIANMTTEW GALSGLFPVDKTLIDWYKNRLQKLGTNNHPRINPKTIRALEEKAKIPKADKDAHYAKKLI IDLATLTHYVSGPNSVKVSNTVQDLSQQDIKINKAYLVSCTNSRLSDLQSAADVVCPTGD LNKVNKVAPGVEFYVAAASSEIEADARKSGAWEKLLKAGCIPLPSGCGPCIGLGAGLLEP GEVGISATNRNFKGRMGSKDALAYLASPAVVAASAVLGKISSPAEVLSTSEIPFSGVKTE IIENPVVEEEVNAQTEAPKQSVEILEGFPREFSGELVLCDADNINTDGIYPGKYTYQDDV PKEKMAQVCMENYDAEFRTKVHPGDIVVSGFNFGTGSSREQAATALLAKGINLVVSGSFG NIFSRNSINNALLTLEIPALIKKLREKYQGAPKELTRRTGWFLKWDVADAKVVVTEGSLD GPVILEQKVGELGKNLQEIIVKGGLEGWVKSQL | ||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||
| External Links |
| ||||||||||||||||||||||||||||||
| General Reference |
| ||||||||||||||||||||||||||||||