Leucine Degradation
| Source | ID | Pathway |
|---|---|---|
| PathWhiz | PW002490 |
Pathway Metabolites
| YMDB ID | Name CAS Number | IUPAC Name | Formula Weight | Structure |
|---|---|---|---|---|
| YMDB00110 | NAD 53-84-9 | 1-[(2R,3R,4S,5R)-5-[({[({[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy](hydroxy)phosphoryl}oxy)methyl]-3,4-dihydroxyoxolan-2-yl]-3-carbamoyl-1lambda5-pyridin-1-ylium | C21H28N7O14P2 664.433 | 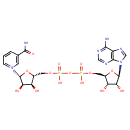 |
| YMDB00143 | NADH 58-68-4 | [({[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy]({[(2R,3S,4R,5R)-5-(3-carbamoyl-1,4-dihydropyridin-1-yl)-3,4-dihydroxyoxolan-2-yl]methoxy})phosphinic acid | C21H29N7O14P2 665.441 | 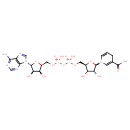 |
| YMDB00153 | Oxoglutaric acid 328-50-7 | 2-oxopentanedioic acid | C5H6O5 146.0981 | 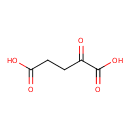 |
| YMDB00271 | L-Glutamic acid 56-86-0 | (2S)-2-aminopentanedioic acid | C5H9NO4 147.1293 | 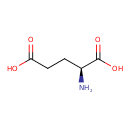 |
| YMDB00387 | L-Leucine 61-90-5 | (2S)-2-amino-4-methylpentanoic acid | C6H13NO2 131.1729 |  |
| YMDB00388 | Ketoleucine 816-66-0 | 4-methyl-2-oxopentanoic acid | C6H10O3 130.1418 |  |
| YMDB00499 | 3-methylbutanal 590-86-3 | 3-methylbutanal | C5H10O 86.1323 |  |
| YMDB00570 | isoamylol 123-51-3 | 3-methylbutan-1-ol | C5H12O 88.1482 |  |
| YMDB00862 | hydron 12408-02-5 | hydron | H 1.00794 |  |
| YMDB00912 | Carbon dioxide 124-38-9 | methanedione | CO2 44.0095 | 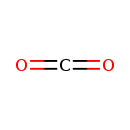 |
Pathway Proteins
| Uniprot ID | Gene Name Locus | Name | Type | Metabolites |
|---|---|---|---|---|
| P47176 | BAT2 YJR148W | Branched-chain-amino-acid aminotransferase, cytosolic | Enzyme | |
| P38891 | BAT1 YHR208W | Branched-chain-amino-acid aminotransferase, mitochondrial | Enzyme | |
| P25377 | ADH7 YCR105W | NADP-dependent alcohol dehydrogenase 7 | Enzyme | |
| Q06408 | ARO10 YDR380W | Transaminated amino acid decarboxylase | Enzyme | |
| Q04894 | ADH6 YMR318C | NADP-dependent alcohol dehydrogenase 6 | Enzyme | |
| Q07471 | THI3 YDL080C | Thiamine metabolism regulatory protein THI3 | Enzyme |