| Identification |
|---|
| YMDB ID | YMDB15900 |
|---|
| Name | PE-NMe(20:4(5Z,8Z,11Z,14Z)/20:4(5Z,8Z,11Z,14Z)) |
|---|
| Species | Saccharomyces cerevisiae |
|---|
| Strain | Brewer's yeast |
|---|
| Description | PE-NMe(20:4(5Z,8Z,11Z,14Z)/20:4(5Z,8Z,11Z,14Z)) is a monomethylphosphatidylethanolamine. It is a glycerophospholipid, and is formed by sequential methylation of phosphatidylethanolamine as part of a mechanism for biosynthesis of phosphatidylcholine. Monomethylphosphatidylethanolamines are usually found at trace levels in animal or plant tissues. They can have many different combinations of fatty acids of varying lengths and saturation attached at the C-1 and C-2 positions. PE-NMe(20:4(5Z,8Z,11Z,14Z)/20:4(5Z,8Z,11Z,14Z)), in particular, consists of two 5Z,8Z,11Z,14Z-eicosatetraenoyl chain at positions C-1 and C2. Fatty acids containing 16, 18 and 20 carbons are the most common. Phospholipids, are ubiquitous in nature and are key components of the lipid bilayer of cells, as well as being involved in metabolism and signaling. |
|---|
| Structure | |
|---|
| Synonyms | - Monomethylphosphatidylethanolamine
- Monomethylphosphatidylethanolamine(20:4(5Z,8Z,11Z,14Z)/20:4(5Z,8Z,11Z,14Z))
|
|---|
| CAS number | Not Available |
|---|
| Weight | Average: 802.087
Monoisotopic: 801.530855409 |
|---|
| InChI Key | MPTMTOMJDUSOGW-UUYLMBBPSA-N |
|---|
| InChI | InChI=1S/C46H76NO8P/c1-4-6-8-10-12-14-16-18-20-22-24-26-28-30-32-34-36-38-45(48)52-42-44(43-54-56(50,51)53-41-40-47-3)55-46(49)39-37-35-33-31-29-27-25-23-21-19-17-15-13-11-9-7-5-2/h12-15,18-21,24-27,30-33,44,47H,4-11,16-17,22-23,28-29,34-43H2,1-3H3,(H,50,51)/b14-12-,15-13-,20-18-,21-19-,26-24-,27-25-,32-30-,33-31- |
|---|
| IUPAC Name | {2,3-bis[(5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoyloxy]propoxy}[2-(methylamino)ethoxy]phosphinic acid |
|---|
| Traditional IUPAC Name | 2,3-bis[(5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoyloxy]propoxy(2-(methylamino)ethoxy)phosphinic acid |
|---|
| Chemical Formula | C46H76NO8P |
|---|
| SMILES | [H]C(COC(=O)CCC\C=C/C\C=C/C\C=C/C\C=C/CCCCC)(COP(O)(=O)OCCNC)OC(=O)CCC\C=C/C\C=C/C\C=C/C\C=C/CCCCC |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as monomethylphosphatidylethanolamines. These are lipids with a structure containing a glycerol moiety linked at its terminal C3 atom to a N-methylphosphoethanolamine group, and at its C1 and C2 terminal atoms by an acyl group. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Glycerophospholipids |
|---|
| Sub Class | Glycerophosphoethanolamines |
|---|
| Direct Parent | Monomethylphosphatidylethanolamines |
|---|
| Alternative Parents | |
|---|
| Substituents | - Monomethylphosphatidylethanolamine
- Phosphoethanolamine
- Fatty acid ester
- Dialkyl phosphate
- Dicarboxylic acid or derivatives
- Organic phosphoric acid derivative
- Phosphoric acid ester
- Alkyl phosphate
- Fatty acyl
- Amino acid or derivatives
- Carboxylic acid ester
- Secondary amine
- Secondary aliphatic amine
- Carboxylic acid derivative
- Organic oxygen compound
- Organooxygen compound
- Organonitrogen compound
- Amine
- Organic nitrogen compound
- Organopnictogen compound
- Carbonyl group
- Organic oxide
- Hydrocarbon derivative
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Charge | 0 |
|---|
| Melting point | Not Available |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Water Solubility | Not Available | PhysProp | | LogP | Not Available | PhysProp |
|
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations | - Endoplasmic reticulum
- Mitochondria
|
|---|
| Organoleptic Properties | Not Available |
|---|
| SMPDB Pathways | | Lysolipid incorporation into ER PC(20:4(5Z,8Z,11Z,14Z)/20:4(5Z,8Z,11Z,14Z)) | PW002790 | 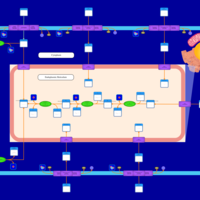 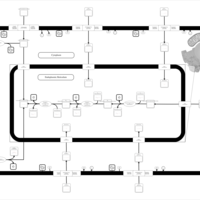 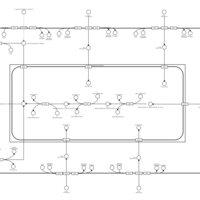 | | Lysolipid incorporation into Mitochondria PC(20:4(5Z,8Z,11Z,14Z)/20:4(5Z,8Z,11Z,14Z)) | PW002798 | 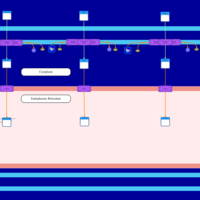 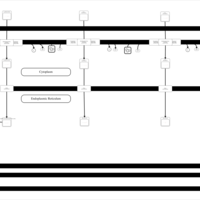 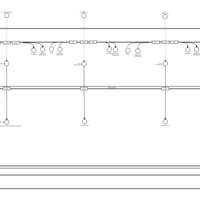 |
|
|---|
| KEGG Pathways | Not Available |
|---|
| SMPDB Reactions | | PE(20:4(5Z,8Z,11Z,14Z)/20:4(5Z,8Z,11Z,14Z))
+
S-Adenosyl-L-methionine
→
PE-NMe(20:4(5Z,8Z,11Z,14Z)/20:4(5Z,8Z,11Z,14Z))
+
hydron
+
S-Adenosylhomocysteine
| | PE-NMe(20:4(5Z,8Z,11Z,14Z)/20:4(5Z,8Z,11Z,14Z))
+
S-Adenosyl-L-methionine
→
PE-NMe2(20:4(5Z,8Z,11Z,14Z)/20:4(5Z,8Z,11Z,14Z))
+
S-Adenosylhomocysteine
+
hydron
|
|
|---|
| KEGG Reactions | Not Available |
|---|
| Concentrations |
|---|
| Intracellular Concentrations | Not Available |
|---|
| Extracellular Concentrations | Not Available |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-4000003790-a58c789c5b523b581cba | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a4i-8300009800-fd4bb8b072fc479f3925 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4i-4931000000-76281ec4184051bb8e55 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udi-2500100290-b7eeb40f22e33b160d2b | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0udj-5529721010-dd04b4be71993a0bd307 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0ufr-9602010000-971005d77ef8af3e194d | JSpectraViewer |
|
|---|
| References |
|---|
| References: | - Rattray JB, Schibeci A, Kidby DK. (1975). "Lipids of yeasts." Bacteriol Rev. 1975 Sep;39(3):197-231.240350
|
|---|
| Synthesis Reference: | Not Available |
|---|
| External Links: | | Resource | Link |
|---|
| CHEBI ID | Not Available | | HMDB ID | HMDB0112976 | | Pubchem Compound ID | 131820091 | | Kegg ID | Not Available | | ChemSpider ID | Not Available | | FOODB ID | Not Available | | Wikipedia ID | Not Available | | BioCyc ID | Not Available |
|
|---|
