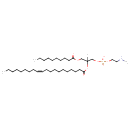
PE-NMe(10:0/20:1(11Z)) (YMDB13873)
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| YMDB ID | YMDB13873 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name | PE-NMe(10:0/20:1(11Z)) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Species | Saccharomyces cerevisiae | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Strain | Brewer's yeast | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | PE-NMe(10:0/20:1(11Z)) is a monomethylphosphatidylethanolamine. It is a glycerophospholipid, and is formed by sequential methylation of phosphatidylethanolamine as part of a mechanism for biosynthesis of phosphatidylcholine. Monomethylphosphatidylethanolamines are usually found at trace levels in animal or plant tissues. They can have many different combinations of fatty acids of varying lengths and saturation attached at the C-1 and C-2 positions. PE-NMe(10:0/20:1(11Z)), in particular, consists of one decanoyl chain to the C-1 atom, and one 11Z-eicosenoyl to the C-2 atom. Fatty acids containing 16, 18 and 20 carbons are the most common. Phospholipids, are ubiquitous in nature and are key components of the lipid bilayer of cells, as well as being involved in metabolism and signaling. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Weight | Average: 675.929 Monoisotopic: 675.483905216 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key | RWRCMLNYPHYBEK-NXVVXOECSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI | InChI=1S/C36H70NO8P/c1-4-6-8-10-12-13-14-15-16-17-18-19-20-21-23-25-27-29-36(39)45-34(33-44-46(40,41)43-31-30-37-3)32-42-35(38)28-26-24-22-11-9-7-5-2/h15-16,34,37H,4-14,17-33H2,1-3H3,(H,40,41)/b16-15- | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name | [3-(decanoyloxy)-2-[(11Z)-icos-11-enoyloxy]propoxy][2-(methylamino)ethoxy]phosphinic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name | 3-(decanoyloxy)-2-[(11Z)-icos-11-enoyloxy]propoxy(2-(methylamino)ethoxy)phosphinic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula | C36H70NO8P | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES | [H]C(COC(=O)CCCCCCCCC)(COP(O)(=O)OCCNC)OC(=O)CCCCCCCCC\C=C/CCCCCCCC | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge | 0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Organoleptic Properties | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMPDB Pathways |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG Pathways | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMPDB Reactions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG Reactions | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Concentrations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Intracellular Concentrations | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Extracellular Concentrations | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in phosphatidylethanolamine N-methyltransferas
- Specific function:
- S-adenosyl-L-methionine + phosphatidylethanolamine = S-adenosyl-L-homocysteine + phosphatidyl-N-methylethanolamine
- Gene Name:
- PEM1
- Uniprot ID:
- P05374
- Molecular weight:
- 101203.0
Reactions
| S-adenosyl-L-methionine + phosphatidylethanolamine → S-adenosyl-L-homocysteine + phosphatidyl-N-methylethanolamine. |
- General function:
- Involved in N-methyltransferase activity
- Specific function:
- S-adenosyl-L-methionine + phospholipid olefinic fatty acid = S-adenosyl-L-homocysteine + phospholipid methylene fatty acid
- Gene Name:
- OPI3
- Uniprot ID:
- P05375
- Molecular weight:
- 23150.09961
Reactions
| S-adenosyl-L-methionine + phospholipid olefinic fatty acid → S-adenosyl-L-homocysteine + phospholipid methylene fatty acid. |