| Identification |
|---|
| YMDB ID | YMDB01493 |
|---|
| Name | Ubiquinone 6 |
|---|
| Species | Saccharomyces cerevisiae |
|---|
| Strain | Brewer's yeast |
|---|
| Description | Ubiquinone 6 belongs to the class of organic compounds known as ubiquinones. These are coenzyme Q derivatives containing a 5, 6-dimethoxy-3-methyl(1,4-benzoquinone) moiety to which an isoprenyl group is attached at ring position 2(or 6). Based on a literature review a significant number of articles have been published on Ubiquinone 6. |
|---|
| Structure | |
|---|
| Synonyms | - (all-E)-2-(3,7,11,15,19,23-hexamethyl-2,6,10,14,18,22-tetracosahexaenyl)-5,6-dimethoxy-3-methyl-2,5-cyclohexadiene-1,4-dione
- 2,3-dimethoxy-5-methyl-6-hexaprenyl-1,4-benzoquinone
- Coenzyme Q6
- Coenzyme Qq6
- CoQ6
- Ubiquinone 30
- Ubiquinone 6
- Ubiquinone Q6
- Ubiquinone-6
- Coenzyme-Q6
- CoQ-6
- Ubiquinone(6)
|
|---|
| CAS number | 1065-31-2 |
|---|
| Weight | Average: 590.8754
Monoisotopic: 590.433510344 |
|---|
| InChI Key | GXNFPEOUKFOTKY-LPHQIWJTSA-N |
|---|
| InChI | InChI=1S/C39H58O4/c1-28(2)16-11-17-29(3)18-12-19-30(4)20-13-21-31(5)22-14-23-32(6)24-15-25-33(7)26-27-35-34(8)36(40)38(42-9)39(43-10)37(35)41/h16,18,20,22,24,26H,11-15,17,19,21,23,25,27H2,1-10H3/b29-18+,30-20+,31-22+,32-24+,33-26+ |
|---|
| IUPAC Name | 2-(3,7,11,15,19,23-hexamethyltetracosa-2,6,10,14,18,22-hexaen-1-yl)-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione |
|---|
| Traditional IUPAC Name | 2-(3,7,11,15,19,23-hexamethyltetracosa-2,6,10,14,18,22-hexaen-1-yl)-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione |
|---|
| Chemical Formula | C39H58O4 |
|---|
| SMILES | COC1=C(OC)C(=O)C(C\C=C(/C)CC\C=C(/C)CC\C=C(/C)CC\C=C(/C)CC\C=C(/C)CCC=C(C)C)=C(C)C1=O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as ubiquinones. These are coenzyme Q derivatives containing a 5, 6-dimethoxy-3-methyl(1,4-benzoquinone) moiety to which an isoprenyl group is attached at ring position 2(or 6). |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Quinone and hydroquinone lipids |
|---|
| Direct Parent | Ubiquinones |
|---|
| Alternative Parents | |
|---|
| Substituents | - Polyprenylbenzoquinone
- Sesterterpenoid
- Ubiquinone skeleton
- Quinone
- P-benzoquinone
- Vinylogous ester
- Cyclic ketone
- Ketone
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Carbonyl group
- Aliphatic homomonocyclic compound
|
|---|
| Molecular Framework | Aliphatic homomonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State | Liquid |
|---|
| Charge | 0 |
|---|
| Melting point | Not Available |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Water Solubility | Not Available | PhysProp | | LogP | Not Available | PhysProp |
|
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations | Not Available |
|---|
| Organoleptic Properties | Not Available |
|---|
| SMPDB Pathways | |
|---|
| KEGG Pathways | | Ubiquinone and other terpenoid-quinone biosynthesis | ec00130 | 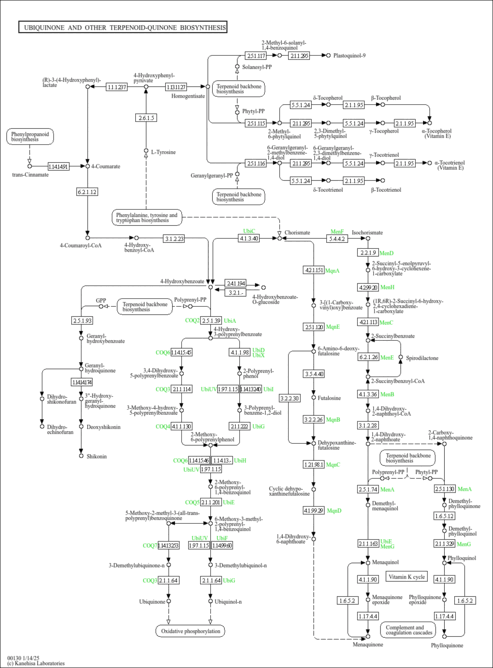 |
|
|---|
| SMPDB Reactions | |
|---|
| KEGG Reactions | Not Available |
|---|
| Concentrations |
|---|
| Intracellular Concentrations | Not Available |
|---|
| Extracellular Concentrations | Not Available |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0fi9-2268490000-d4a46fb7bbfe567b3bc9 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0007-0112190000-0e3f9b8afdf1ec72101d | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-002b-0649330000-25374eb6b7e2528209f9 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00kb-2125910000-2353dc21b36ad0226438 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000i-0000090000-888a7e1c1887cd6289a7 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-000i-0000090000-cdb5f8e03d488aabf72f | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0079-9100580000-0ddcd7334398c6806be9 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0007-0212190000-d6790c276a238e59aeca | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-002b-0649330000-25374eb6b7e2528209f9 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00mk-2125910000-fd96f37265811c6c3531 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000i-0000090000-1e48e9465d0a2b5e046c | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-000i-0100090000-308b58d9d2d89e2a0150 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0079-8100590000-fa8ea854e31b895398f6 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000e-4619380000-15b4ea76379fc8709a54 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0002-4915010000-9f9f91ee19707f8366d2 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000y-8924000000-472232f8797340057410 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000i-0000090000-652b47e044f4b53fb940 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-07br-0611090000-96454b02ed13c80d5269 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-003r-1694370000-5e9b03bff5e8c4cb6b63 | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer |
|
|---|
| References |
|---|
| References: | - Luttik, M. A., Overkamp, K. M., Kotter, P., de Vries, S., van Dijken, J. P., Pronk, J. T. (1998). "The Saccharomyces cerevisiae NDE1 and NDE2 genes encode separate mitochondrial NADH dehydrogenases catalyzing the oxidation of cytosolic NADH." J Biol Chem 273:24529-24534.9733747
- Yannai S. Dictionary of food compounds with CD-ROM: additives, flavors, and ingredients. Chapman & Hall/CRC; 2004.
|
|---|
| Synthesis Reference: | Not Available |
|---|
| External Links: | |
|---|

