| Identification |
|---|
| YMDB ID | YMDB00164 |
|---|
| Name | Thiamine monophosphate |
|---|
| Species | Saccharomyces cerevisiae |
|---|
| Strain | Baker's yeast |
|---|
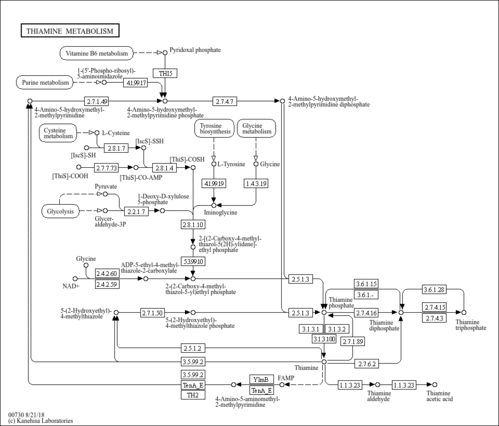
| Description | Thiamine monophosphate, also known as thiamin phosphoric acid or monophosphothiaminum, belongs to the class of organic compounds known as thiamine phosphates. These are thiamine derivatives in which the hydroxyl group of the ethanol moiety is substituted by a phosphate group. Thiamine monophosphate is a very strong basic compound (based on its pKa). Thiamine monophosphate exists in all eukaryotes, ranging from yeast to humans. Within yeast, thiamine monophosphate participates in a number of enzymatic reactions. In particular, thiamine monophosphate can be biosynthesized from 2-methyl-4-amino-5-hydroxymethylpyrimidine diphosphate and 4-methyl-5-(2-phosphonooxyethyl)thiazole through the action of the enzyme thiamine biosynthetic bifunctional enzyme. In addition, thiamine monophosphate can be converted into thiamine through its interaction with the enzyme acid phosphatases. In yeast, thiamine monophosphate is involved in the metabolic pathway called the vitamin b1/thiamine metabolism pathway. |
|---|
| Structure | |
|---|
| Synonyms | - Aneurin-monophosphorsaeureester
- Aneurine monophosphate
- Monofosfotiamina
- Monophosphoric ester of thiamine
- Monophosphothiamine
- Monophosphothiaminum
- Phosphothiaminum
- Thiamin dihydrogenphosphate
- thiamin monophosphate
- Thiamin phosphate
- Thiamine monophosphate
- Thiamine monophosphate dihydrate
- Thiamine monophosphic acid
- thiamine phosphate
- Tiamina monofosfato
- TMP
- tmp, thiamin phosphate
- THIAMIN phosphoric acid
- Thiamine monophosphoric acid
- monoPhosphate, thiamine
- Phosphoester, thiamine
- Thiamine phosphoester
|
|---|
| CAS number | 495-23-8 |
|---|
| Weight | Average: 344.327
Monoisotopic: 344.070812254 |
|---|
| InChI Key | HZSAJDVWZRBGIF-UHFFFAOYSA-N |
|---|
| InChI | InChI=1S/C12H17N4O4PS/c1-8-11(3-4-20-21(17,18)19)22-7-16(8)6-10-5-14-9(2)15-12(10)13/h5,7H,3-4,6H2,1-2H3,(H3-,13,14,15,17,18,19) |
|---|
| IUPAC Name | 3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-[2-(hydrogen phosphonooxy)ethyl]-4-methyl-1,3-thiazol-3-ium |
|---|
| Traditional IUPAC Name | 3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-[2-(hydrogen phosphonooxy)ethyl]-4-methyl-1,3-thiazol-3-ium |
|---|
| Chemical Formula | C12H17N4O4PS |
|---|
| SMILES | [H]OP([O-])(=O)OC([H])([H])C([H])([H])C1=C([N+](=C([H])S1)C([H])([H])C1=C([H])N=C(N=C1N([H])[H])C([H])([H])[H])C([H])([H])[H] |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as thiamine phosphates. These are thiamine derivatives in which the hydroxyl group of the ethanol moiety is substituted by a phosphate group. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Diazines |
|---|
| Sub Class | Pyrimidines and pyrimidine derivatives |
|---|
| Direct Parent | Thiamine phosphates |
|---|
| Alternative Parents | |
|---|
| Substituents | - Thiamine-phosphate
- 4,5-disubstituted 1,3-thiazole
- Aminopyrimidine
- Organic phosphoric acid derivative
- Phosphoric acid ester
- Alkyl phosphate
- Imidolactam
- Azole
- Thiazole
- Heteroaromatic compound
- Azacycle
- Amine
- Hydrocarbon derivative
- Primary amine
- Organic oxide
- Organooxygen compound
- Organonitrogen compound
- Organopnictogen compound
- Organic nitrogen compound
- Organic oxygen compound
- Organic zwitterion
- Aromatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aromatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Charge | 0 |
|---|
| Melting point | Not Available |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Water Solubility | Not Available | PhysProp | | LogP | Not Available | PhysProp |
|
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations | |
|---|
| Organoleptic Properties | Not Available |
|---|
| SMPDB Pathways | |
|---|
| KEGG Pathways | |
|---|
| SMPDB Reactions | |
|---|
| KEGG Reactions | |
|---|
| Concentrations |
|---|
| Intracellular Concentrations | Not Available |
|---|
| Extracellular Concentrations | Not Available |
|---|
| Spectra |
|---|
| Spectra | |
|---|
| References |
|---|
| References: | - UniProt Consortium (2011). "Ongoing and future developments at the Universal Protein Resource." Nucleic Acids Res 39:D214-D219.21051339
- Onozuka, M., Konno, H., Kawasaki, Y., Akaji, K., Nosaka, K. (2008). "Involvement of thiaminase II encoded by the THI20 gene in thiamin salvage of Saccharomyces cerevisiae." FEMS Yeast Res 8:266-275.18028398
- Herrgard, M. J., Swainston, N., Dobson, P., Dunn, W. B., Arga, K. Y., Arvas, M., Bluthgen, N., Borger, S., Costenoble, R., Heinemann, M., Hucka, M., Le Novere, N., Li, P., Liebermeister, W., Mo, M. L., Oliveira, A. P., Petranovic, D., Pettifer, S., Simeonidis, E., Smallbone, K., Spasic, I., Weichart, D., Brent, R., Broomhead, D. S., Westerhoff, H. V., Kirdar, B., Penttila, M., Klipp, E., Palsson, B. O., Sauer, U., Oliver, S. G., Mendes, P., Nielsen, J., Kell, D. B. (2008). "A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology." Nat Biotechnol 26:1155-1160.18846089
|
|---|
| Synthesis Reference: | Leder, Irwin G. Enzymic synthesis of thiamine monophosphate. Journal of Biological Chemistry (1961), 236 3066-71. |
|---|
| External Links: | |
|---|