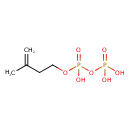| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-002b-9600000000-930df1e6e1c7e4eed108 | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | JSpectraViewer | | LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 0V, negative | splash10-004i-0190000000-d2895fc0c10b183f43ee | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 1V, negative | splash10-056r-3980000000-627cfd435c85e88822b5 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 1V, negative | splash10-056r-3980000000-23fe88b159964959e66f | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 5V, negative | splash10-056r-2960000000-19354ef717c66764cd68 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 8V, negative | splash10-056r-9740000000-022ac5cb9631af8164ee | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 10V, negative | splash10-004i-9300000000-aee23682a2c1fa358b3b | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 11V, negative | splash10-004i-9100000000-6318f24bf41c952270c7 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 14V, negative | splash10-004i-9000000000-84d7ad80acc22ebadc08 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 16V, negative | splash10-004i-9000000000-52e623d1c0e3c5eec628 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 19V, negative | splash10-004i-9000000000-71f08ee06355626ebfd6 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - n/a 17V, negative | splash10-0a4i-0900000000-8b768e6e6fd82a9e52fc | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - n/a 17V, negative | splash10-004i-9000000000-5ef060d5ec4b80c35c1a | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - n/a 17V, negative | splash10-00kr-0900000000-1e624fb025fa3835685b | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - n/a 17V, negative | splash10-014i-1900000000-2c64756ee3a8e3d9493c | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 1V, negative | splash10-0006-0090000000-5185751820ffb8d752a3 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 3V, negative | splash10-0006-0090000000-03743901b1bb8cf365e9 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 4V, negative | splash10-0006-2090000000-9e4213acb94b27ef7c83 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 5V, negative | splash10-004l-9080000000-43b0fb277b1db3562d57 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 7V, negative | splash10-004i-9010000000-863fed72efb5d1767ed2 | JSpectraViewer | MoNA | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014j-9670000000-fa3d42a8ff93d345cea7 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-014i-9200000000-78e51dcb039157700c59 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-014i-9100000000-a585b43371cb54efe2eb | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-0490000000-391e90cfd97e6adcbb06 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9510000000-fed3169adff6f1179e01 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-5c099085cd907446a2fb | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer |
|
|---|
| References: | - UniProt Consortium (2011). "Ongoing and future developments at the Universal Protein Resource." Nucleic Acids Res 39:D214-D219.21051339
- Scheer, M., Grote, A., Chang, A., Schomburg, I., Munaretto, C., Rother, M., Sohngen, C., Stelzer, M., Thiele, J., Schomburg, D. (2011). "BRENDA, the enzyme information system in 2011." Nucleic Acids Res 39:D670-D676.21062828
- Herrgard, M. J., Swainston, N., Dobson, P., Dunn, W. B., Arga, K. Y., Arvas, M., Bluthgen, N., Borger, S., Costenoble, R., Heinemann, M., Hucka, M., Le Novere, N., Li, P., Liebermeister, W., Mo, M. L., Oliveira, A. P., Petranovic, D., Pettifer, S., Simeonidis, E., Smallbone, K., Spasic, I., Weichart, D., Brent, R., Broomhead, D. S., Westerhoff, H. V., Kirdar, B., Penttila, M., Klipp, E., Palsson, B. O., Sauer, U., Oliver, S. G., Mendes, P., Nielsen, J., Kell, D. B. (2008). "A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology." Nat Biotechnol 26:1155-1160.18846089
- Sato, M., Sato, K., Nishikawa, S., Hirata, A., Kato, J., Nakano, A. (1999). "The yeast RER2 gene, identified by endoplasmic reticulum protein localization mutations, encodes cis-prenyltransferase, a key enzyme in dolichol synthesis." Mol Cell Biol 19:471-483.9858571
- Grabinska, K., Palamarczyk, G. (2002). "Dolichol biosynthesis in the yeast Saccharomyces cerevisiae: an insight into the regulatory role of farnesyl diphosphate synthase." FEMS Yeast Res 2:259-265.12702274
- Hsu, A. Y., Do, T. Q., Lee, P. T., Clarke, C. F. (2000). "Genetic evidence for a multi-subunit complex in the O-methyltransferase steps of coenzyme Q biosynthesis." Biochim Biophys Acta 1484:287-297.10760477
|
|---|