| Identification |
|---|
| YMDB ID | YMDB14943 |
|---|
| Name | PG(16:0/18:0) |
|---|
| Species | Saccharomyces cerevisiae |
|---|
| Strain | Brewer's yeast |
|---|
| Description | PG(16:0/18:0) is a phosphatidylglycerol. Phosphatidylglycerols consist of a glycerol 3-phosphate backbone esterified to either saturated or unsaturated fatty acids on carbons 1 and 2. As is the case with diacylglycerols, phosphatidylglycerols can have many different combinations of fatty acids of varying lengths and saturation attached to the C-1 and C-2 positions. PG(16:0/18:0), in particular, consists of one hexadecanoyl chain to the C-1 atom, and one octadecanoyl to the C-2 atom. In E. coli glycerophospholipid metabolism, phosphatidylglycerol is formed from phosphatidic acid (1,2-diacyl-sn-glycerol 3-phosphate) by a sequence of enzymatic reactions that proceeds via two intermediates, cytidine diphosphate diacylglycerol (CDP-diacylglycerol) and phosphatidylglycerophosphate (PGP, a phosphorylated phosphatidylglycerol). Phosphatidylglycerols, along with CDP-diacylglycerol, also serve as precursor molecules for the synthesis of cardiolipin, a phospholipid found in membranes. |
|---|
| Structure | |
|---|
| Synonyms | - 1-Palmitoyl-2-stearoyl-sn-glycero-3-phospho-(1'-sn-glycerol)
- 1-Palmitoyl-2-stearoyl-sn-glycero-3-phosphoglycerol
- GPG(16:0/18:0)
- GPG(34:0)
- PG(34:0)
- Phosphatidylglycerol(16:0/18:0)
- Phosphatidylglycerol(34:0)
- 1-hexadecanoyl-2-octadecanoyl-sn-glycero-3-phospho-(1'-glycerol)
- PG(16:0/18:0)
- 1-hexadecanoyl-2-octadecanoyl-sn-glycero-3-phosphoglycerol
|
|---|
| CAS number | Not Available |
|---|
| Weight | Average: 751.023
Monoisotopic: 750.54108526 |
|---|
| InChI Key | KBPVYRBBONZJHF-QPPIDDCLSA-N |
|---|
| InChI | InChI=1S/C40H79O10P/c1-3-5-7-9-11-13-15-17-18-20-22-24-26-28-30-32-40(44)50-38(36-49-51(45,46)48-34-37(42)33-41)35-47-39(43)31-29-27-25-23-21-19-16-14-12-10-8-6-4-2/h37-38,41-42H,3-36H2,1-2H3,(H,45,46)/t37-,38+/m0/s1 |
|---|
| IUPAC Name | [(2S)-2,3-dihydroxypropoxy][(2R)-3-(hexadecanoyloxy)-2-(octadecanoyloxy)propoxy]phosphinic acid |
|---|
| Traditional IUPAC Name | (2S)-2,3-dihydroxypropoxy((2R)-3-(hexadecanoyloxy)-2-(octadecanoyloxy)propoxy)phosphinic acid |
|---|
| Chemical Formula | C40H79O10P |
|---|
| SMILES | [H][C@](O)(CO)COP(O)(=O)OC[C@@]([H])(COC(=O)CCCCCCCCCCCCCCC)OC(=O)CCCCCCCCCCCCCCCCC |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as phosphatidylglycerols. These are glycerophosphoglycerols in which two fatty acids are bonded to the 1-glycerol moiety through ester linkages. As is the case with diacylglycerols, phosphatidylglycerols can have many different combinations of fatty acids of varying lengths and saturation attached to the C-1 and C-2 positions. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Glycerophospholipids |
|---|
| Sub Class | Glycerophosphoglycerols |
|---|
| Direct Parent | Phosphatidylglycerols |
|---|
| Alternative Parents | |
|---|
| Substituents | - 1,2-diacylglycerophosphoglycerol
- Fatty acid ester
- Dialkyl phosphate
- Dicarboxylic acid or derivatives
- Organic phosphoric acid derivative
- Phosphoric acid ester
- Alkyl phosphate
- Fatty acyl
- 1,2-diol
- Carboxylic acid ester
- Secondary alcohol
- Carboxylic acid derivative
- Organic oxide
- Organooxygen compound
- Alcohol
- Organic oxygen compound
- Primary alcohol
- Carbonyl group
- Hydrocarbon derivative
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Charge | 0 |
|---|
| Melting point | Not Available |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Water Solubility | Not Available | PhysProp | | LogP | Not Available | PhysProp |
|
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations | |
|---|
| Organoleptic Properties | Not Available |
|---|
| SMPDB Pathways | | Cardiolipin Biosynthesis CL(16:0/18:0/16:0/18:0) | PW012584 |  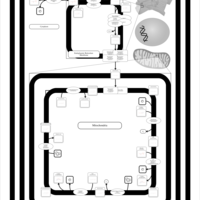 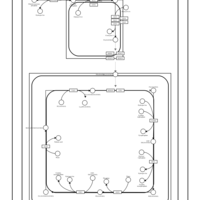 | | Cardiolipin Biosynthesis CL(16:0/18:0/18:0/18:0) | PW012585 | 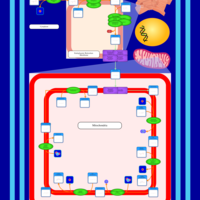 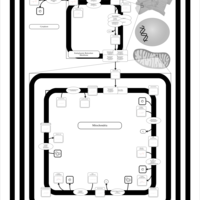  | | Cardiolipin Biosynthesis CL(16:0/18:0/18:0/18:1(11Z)) | PW012586 | 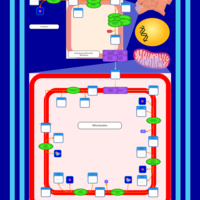 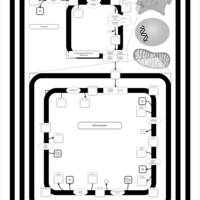 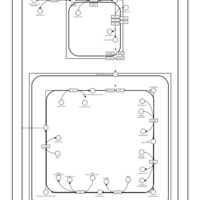 | | Cardiolipin Biosynthesis CL(16:0/18:0/18:0/18:1(9Z)) | PW012587 | 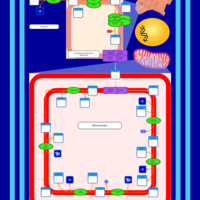   | | Cardiolipin Biosynthesis CL(16:0/18:0/18:0/20:0) | PW012588 |    |
|
|---|
| KEGG Pathways | Not Available |
|---|
| SMPDB Reactions |
|
|---|
| KEGG Reactions | Not Available |
|---|
| Concentrations |
|---|
| Intracellular Concentrations | Not Available |
|---|
| Extracellular Concentrations | Not Available |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_2) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_3) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0aps-0190200200-fe1f1909e577bf28e2ae | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a70-4290100000-3498d14b376929615de5 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9030100000-f846283ee757359023a5 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-0000000900-01f8acb19a844bd6d2dd | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-053s-0090300400-7926d39d0fa0623a46d1 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-053s-0190300400-98baa113a06477af5f07 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0gei-3190510500-2c4c5fb0d7aa1c7a97f0 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00or-4291311100-589d9cd95c0d05ffeaf7 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0bvi-8192021000-e57c6036faab210bfa03 | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer |
|
|---|
| References |
|---|
| References: | - Rattray JB, Schibeci A, Kidby DK. (1975). "Lipids of yeasts." Bacteriol Rev. 1975 Sep;39(3):197-231.240350
|
|---|
| Synthesis Reference: | Not Available |
|---|
| External Links: | |
|---|
