| Identification |
|---|
| YMDB ID | YMDB14233 |
|---|
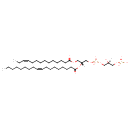
| Name | PGP(14:1(11Z)/18:1(9Z)) |
|---|
| Species | Saccharomyces cerevisiae |
|---|
| Strain | Brewer's yeast |
|---|
| Description | PGP(14:1(11Z)/18:1(9Z)) belongs to the class of glycerophosphoglycerophosphates, also called phosphatidylglycerophosphates (PGPs). These lipids contain a common glycerophosphate skeleton linked to at least one fatty acyl chain and a glycero-3-phosphate moiety. As is the case with diacylglycerols, phosphatidylglycerophosphates can have many different combinations of fatty acids of varying lengths and saturation attached to the C-1 and C-2 positions. PGP(14:1(11Z)/18:1(9Z)), in particular, consists of one 11Z-tetradecenoyl chain to the C-1 atom, and one 9Z-octadecenoyl to the C-2 atom. In E. coli, PGPs can be found in the cytoplasmic membrane. The are synthesized by the addition of glycerol 3-phosphate to a CDP-diacylglycerol. In turn, PGPs are dephosphorylated to Phosphatidylglycerols (PGs) by the enzyme Phosphatidylglycerophosphatase. |
|---|
| Structure | |
|---|
| Synonyms | Not Available |
|---|
| CAS number | Not Available |
|---|
| Weight | Average: 798.929
Monoisotopic: 798.444816374 |
|---|
| InChI Key | HTJFSIVRMAPSHX-WKPQWOHKSA-N |
|---|
| InChI | InChI=1S/C38H72O13P2/c1-3-5-7-9-11-13-15-16-17-18-20-22-24-26-28-30-38(41)51-36(34-50-53(45,46)49-32-35(39)31-48-52(42,43)44)33-47-37(40)29-27-25-23-21-19-14-12-10-8-6-4-2/h6,8,16-17,35-36,39H,3-5,7,9-15,18-34H2,1-2H3,(H,45,46)(H2,42,43,44)/b8-6-,17-16-/t35-,36+/m0/s1 |
|---|
| IUPAC Name | [(2S)-2-hydroxy-3-({hydroxy[(2R)-2-[(9Z)-octadec-9-enoyloxy]-3-[(11Z)-tetradec-11-enoyloxy]propoxy]phosphoryl}oxy)propoxy]phosphonic acid |
|---|
| Traditional IUPAC Name | (2S)-2-hydroxy-3-{[hydroxy((2R)-2-[(9Z)-octadec-9-enoyloxy]-3-[(11Z)-tetradec-11-enoyloxy]propoxy)phosphoryl]oxy}propoxyphosphonic acid |
|---|
| Chemical Formula | C38H72O13P2 |
|---|
| SMILES | [H][C@](O)(COP(O)(O)=O)COP(O)(=O)OC[C@@]([H])(COC(=O)CCCCCCCCC\C=C/CC)OC(=O)CCCCCCC\C=C/CCCCCCCC |
|---|
| Chemical Taxonomy |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Charge | 0 |
|---|
| Melting point | Not Available |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Water Solubility | Not Available | PhysProp | | LogP | Not Available | PhysProp |
|
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations | |
|---|
| Organoleptic Properties | Not Available |
|---|
| SMPDB Pathways | | Cardiolipin Biosynthesis CL(14:1(11Z)/18:1(9Z)/14:1(11Z)/18:1(11Z)) | PW010296 |    | | Cardiolipin Biosynthesis CL(14:1(11Z)/18:1(9Z)/14:1(11Z)/18:1(9Z)) | PW010297 |    | | Cardiolipin Biosynthesis CL(14:1(11Z)/18:1(9Z)/14:1(9Z)/18:1(11Z)) | PW010298 |    | | Cardiolipin Biosynthesis CL(14:1(11Z)/18:1(9Z)/14:1(9Z)/18:1(9Z)) | PW010299 |    | | Cardiolipin Biosynthesis CL(14:1(11Z)/18:1(9Z)/18:1(11Z)/18:1(11Z)) | PW010300 |    |
|
|---|
| KEGG Pathways | Not Available |
|---|
| SMPDB Reactions | |
|---|
| KEGG Reactions | Not Available |
|---|
| Concentrations |
|---|
| Intracellular Concentrations | Not Available |
|---|
| Extracellular Concentrations | Not Available |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-067j-1390051600-ada38af7b63c681d36ac | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0apj-2490022200-8eb7c781c8b570ede46e | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4i-5491021000-916292a3fd0bf488dbca | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0059-3090020200-8479550b654a546330ab | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9050000000-907fd19c6932ea7d45bd | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9010000000-d1429230bb9d13bb15cc | JSpectraViewer |
|
|---|
| References |
|---|
| References: | - Rattray JB, Schibeci A, Kidby DK. (1975). "Lipids of yeasts." Bacteriol Rev. 1975 Sep;39(3):197-231.240350
|
|---|
| Synthesis Reference: | Not Available |
|---|
| External Links: | | Resource | Link |
|---|
| CHEBI ID | Not Available | | HMDB ID | Not Available | | Pubchem Compound ID | Not Available | | Kegg ID | Not Available | | ChemSpider ID | Not Available | | FOODB ID | Not Available | | Wikipedia ID | Not Available | | BioCyc ID | Not Available |
|
|---|