| Identification |
|---|
| YMDB ID | YMDB11762 |
|---|
| Name | 2-MLCL(12:0/12:0/14:1(9Z)/0:0) |
|---|
| Species | Saccharomyces cerevisiae |
|---|
| Strain | Brewer's yeast |
|---|
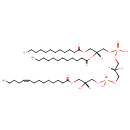
| Description | 2-MLCL(12:0/12:0/14:1(9Z)/0:0) is a monolysocardiolipin (MLCL). Monolysocardiolipins have three fatty acid tails, instead of the usual two. 2-MLCL(12:0/12:0/14:1(9Z)/0:0) contains two chains of dodecanoic acid at the C1 and C2 positions, one chain of (9Z-tetradecenoyl) at the C3 position, one chain of at the C4 position. MLCL is present in eukaryotes as part of the metabolic cycle of mitochondrial lipids. Removal of one acyl chain from a cardiolipin results in generation of monolysocardiolipin (MLCL). MLCL has been used as an intermediate in the synthesis of spin-labeled CL to study the interaction of CL with mitochondrial enzymes. Because a role for MLCL has been suggested in apoptosis, this molecule has been used to study its interaction with various enzymes involved in lipid remodeling and apoptosis. There are two species of monolysocardiolipins, 1-MLCL which is missing a fatty acid in position R1 the and 2-MLCL which is missing a fatty acid in position R4. |
|---|
| Structure | |
|---|
| Synonyms | Not Available |
|---|
| CAS number | Not Available |
|---|
| Weight | Average: 973.169
Monoisotopic: 972.570410814 |
|---|
| InChI Key | BJIHCWYNQPRMQK-CEAWJIDOSA-N |
|---|
| InChI | InChI=1S/C47H90O16P2/c1-4-7-10-13-16-19-20-23-25-27-30-33-45(50)57-36-42(48)37-59-64(53,54)60-38-43(49)39-61-65(55,56)62-41-44(63-47(52)35-32-29-26-22-18-15-12-9-6-3)40-58-46(51)34-31-28-24-21-17-14-11-8-5-2/h13,16,42-44,48-49H,4-12,14-15,17-41H2,1-3H3,(H,53,54)(H,55,56)/b16-13-/t42-,43-,44-/m1/s1 |
|---|
| IUPAC Name | [(2R)-2,3-bis(dodecanoyloxy)propoxy][(2R)-2-hydroxy-3-({hydroxy[(2R)-2-hydroxy-3-[(9Z)-tetradec-9-enoyloxy]propoxy]phosphoryl}oxy)propoxy]phosphinic acid |
|---|
| Traditional IUPAC Name | (2R)-2,3-bis(dodecanoyloxy)propoxy((2R)-2-hydroxy-3-{[hydroxy((2R)-2-hydroxy-3-[(9Z)-tetradec-9-enoyloxy]propoxy)phosphoryl]oxy}propoxy)phosphinic acid |
|---|
| Chemical Formula | C47H90O16P2 |
|---|
| SMILES | [H][C@@](O)(COC(=O)CCCCCCC\C=C/CCCC)COP(O)(=O)OC[C@@]([H])(O)COP(O)(=O)OC[C@@]([H])(COC(=O)CCCCCCCCCCC)OC(=O)CCCCCCCCCCC |
|---|
| Chemical Taxonomy |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Charge | 0 |
|---|
| Melting point | Not Available |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Water Solubility | Not Available | PhysProp | | LogP | Not Available | PhysProp |
|
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations | |
|---|
| Organoleptic Properties | Not Available |
|---|
| SMPDB Pathways | | Cardiolipin Biosynthesis CL(12:0/12:0/14:1(9Z)/22:0) | PW005913 | 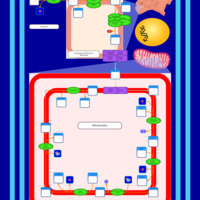 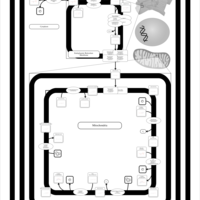 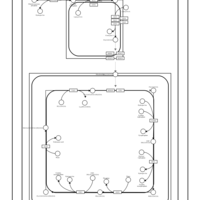 | | Cardiolipin Biosynthesis CL(12:0/12:0/14:1(9Z)/22:1(11Z)) | PW005914 |    | | Cardiolipin Biosynthesis CL(12:0/12:0/14:1(9Z)/22:1(9Z)) | PW005915 |   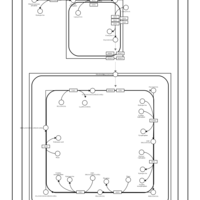 | | Cardiolipin Biosynthesis CL(12:0/12:0/14:1(9Z)/24:0) | PW005916 |    | | Cardiolipin Biosynthesis CL(12:0/12:0/14:1(9Z)/24:1(11Z)) | PW005917 |    |
|
|---|
| KEGG Pathways | Not Available |
|---|
| SMPDB Reactions |
|
|---|
| KEGG Reactions | Not Available |
|---|
| Concentrations |
|---|
| Intracellular Concentrations | Not Available |
|---|
| Extracellular Concentrations | Not Available |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0ac0-0481440809-3d7809c50322b724bf35 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0ac0-1892472505-b082136bf143f3d5253f | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a59-2590320101-ad15629832676d3354af | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-05ea-0891020403-b204d0a8d42131a2bc7f | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-7972020000-f794573f9fbf9c0219a3 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004j-9241010000-9f5a9454a36a75980151 | JSpectraViewer |
|
|---|
| References |
|---|
| References: | - Baile MG, Lu YW, Claypool SM. (2014). "The topology and regulation of cardiolipin biosynthesis and remodeling in yeast." Chem Phys Lipids. 2014 Apr;179:25-31. doi: 10.1016/j.chemphyslip.2013.10.008. Epub 2013 Nov 1.24184646
|
|---|
| Synthesis Reference: | Not Available |
|---|
| External Links: | | Resource | Link |
|---|
| CHEBI ID | Not Available | | HMDB ID | Not Available | | Pubchem Compound ID | Not Available | | Kegg ID | Not Available | | ChemSpider ID | Not Available | | FOODB ID | Not Available | | Wikipedia ID | Not Available | | BioCyc ID | Not Available |
|
|---|