| Identification |
|---|
| YMDB ID | YMDB01211 |
|---|
| Name | PG(16:1(9Z)/16:1(9Z)) |
|---|
| Species | Saccharomyces cerevisiae |
|---|
| Strain | Brewer's yeast |
|---|
| Description | PG(16:1(9Z)/16:1(9Z)) is a phosphatidylglycerol. Phosphatidylglycerols consist of a glycerol 3-phosphate backbone esterified to either saturated or unsaturated fatty acids on carbons 1 and 2. As is the case with diacylglycerols, phosphatidylglycerols can have many different combinations of fatty acids of varying lengths and saturation attached to the C-1 and C-2 positions. PG(16:1(9Z)/16:1(9Z)), in particular, consists of two 9Z-hexadecenoyl chains at positions C-1 and C-2. In E. coli glycerophospholipid metabolism, phosphatidylglycerol is formed from phosphatidic acid (1,2-diacyl-sn-glycerol 3-phosphate) by a sequence of enzymatic reactions that proceeds via two intermediates, cytidine diphosphate diacylglycerol (CDP-diacylglycerol) and phosphatidylglycerophosphate (PGP, a phosphorylated phosphatidylglycerol). Phosphatidylglycerols, along with CDP-diacylglycerol, also serve as precursor molecules for the synthesis of cardiolipin, a phospholipid found in membranes. |
|---|
| Structure | |
|---|
| Synonyms | - 1,2-di(9Z-hexadecenoyl)-rac-glycero-3-phospho-(1'-glycerol)
- 1,2-dipalmitoleoyl-rac-glycero-3-phosphoglycerol
- GPG(16:1/16:1)
- GPG(16:1n7/16:1n7)
- GPG(16:1w7/16:1w7)
- GPG(32:2)
- PG(16:1/16:1)
- PG(16:1n7/16:1n7)
- PG(16:1w7/16:1w7)
- PG(32:2)
- Phosphatidylglycerol(16:1/16:1)
- Phosphatidylglycerol(16:1n7/16:1n7)
- Phosphatidylglycerol(16:1w7/16:1w7)
- Phosphatidylglycerol(32:2)
- 1,2-Di(9Z-hexadecenoyl)-rac-glycero-3-phosphoglycerol
- PG(16:1(9Z)/16:1(9Z))
|
|---|
| CAS number | 66322-31-4 |
|---|
| Weight | Average: 718.95
Monoisotopic: 718.478485484 |
|---|
| InChI Key | GHQNERCMPIDAAU-NWHZUZAFSA-N |
|---|
| InChI | InChI=1S/C38H71O10P/c1-3-5-7-9-11-13-15-17-19-21-23-25-27-29-37(41)45-33-36(34-47-49(43,44)46-32-35(40)31-39)48-38(42)30-28-26-24-22-20-18-16-14-12-10-8-6-4-2/h13-16,35-36,39-40H,3-12,17-34H2,1-2H3,(H,43,44)/b15-13-,16-14-/t35-,36+/m0/s1 |
|---|
| IUPAC Name | [(2R)-2,3-bis[(9Z)-hexadec-9-enoyloxy]propoxy][(2S)-2,3-dihydroxypropoxy]phosphinic acid |
|---|
| Traditional IUPAC Name | (2R)-2,3-bis[(9Z)-hexadec-9-enoyloxy]propoxy((2S)-2,3-dihydroxypropoxy)phosphinic acid |
|---|
| Chemical Formula | C38H71O10P |
|---|
| SMILES | [H][C@](O)(CO)COP(O)(=O)OC[C@@]([H])(COC(=O)CCCCCCC\C=C/CCCCCC)OC(=O)CCCCCCC\C=C/CCCCCC |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as phosphatidylglycerols. These are glycerophosphoglycerols in which two fatty acids are bonded to the 1-glycerol moiety through ester linkages. As is the case with diacylglycerols, phosphatidylglycerols can have many different combinations of fatty acids of varying lengths and saturation attached to the C-1 and C-2 positions. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Glycerophospholipids |
|---|
| Sub Class | Glycerophosphoglycerols |
|---|
| Direct Parent | Phosphatidylglycerols |
|---|
| Alternative Parents | |
|---|
| Substituents | - 1,2-diacylglycerophosphoglycerol
- Fatty acid ester
- Dialkyl phosphate
- Dicarboxylic acid or derivatives
- Organic phosphoric acid derivative
- Phosphoric acid ester
- Alkyl phosphate
- Fatty acyl
- 1,2-diol
- Carboxylic acid ester
- Secondary alcohol
- Carboxylic acid derivative
- Organic oxide
- Organooxygen compound
- Alcohol
- Organic oxygen compound
- Primary alcohol
- Carbonyl group
- Hydrocarbon derivative
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Charge | 0 |
|---|
| Melting point | Not Available |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Water Solubility | Not Available | PhysProp | | LogP | Not Available | PhysProp |
|
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations | Not Available |
|---|
| Organoleptic Properties | Not Available |
|---|
| SMPDB Pathways | | Cardiolipin Biosynthesis CL(16:1(9Z)/16:1(9Z)/16:1(11Z)/16:1(11Z)) | PW012836 | 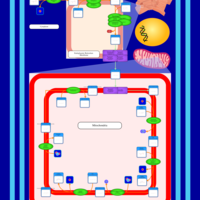 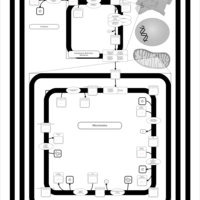 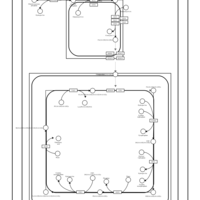 | | Cardiolipin Biosynthesis CL(16:1(9Z)/16:1(9Z)/16:1(11Z)/16:1(9Z)) | PW012837 | 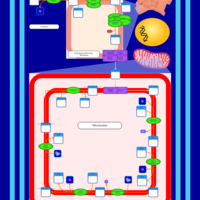 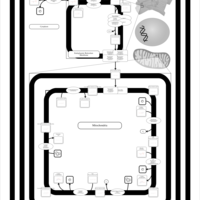 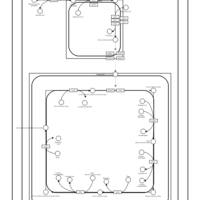 | | Cardiolipin Biosynthesis CL(16:1(9Z)/16:1(9Z)/16:1(11Z)/18:0) | PW012838 | 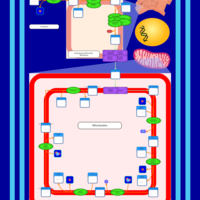 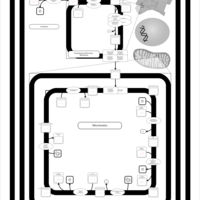 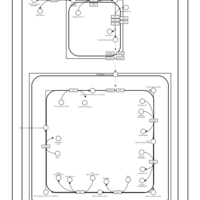 | | Cardiolipin Biosynthesis CL(16:1(9Z)/16:1(9Z)/16:1(11Z)/18:1(11Z)) | PW012839 | 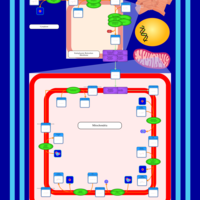 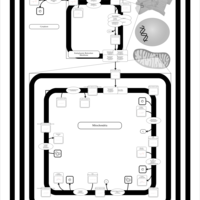 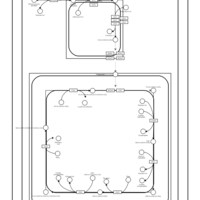 | | Cardiolipin Biosynthesis CL(16:1(9Z)/16:1(9Z)/16:1(11Z)/18:1(9Z)) | PW012840 | 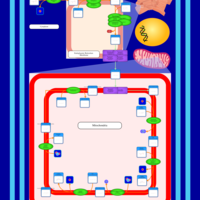 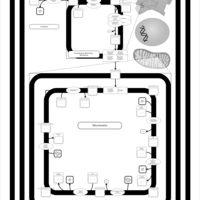  |
|
|---|
| KEGG Pathways | Not Available |
|---|
| SMPDB Reactions |
|
|---|
| KEGG Reactions | Not Available |
|---|
| Concentrations |
|---|
| Intracellular Concentrations | | Intracellular Concentration | Substrate | Growth Conditions | Strain | Citation |
|---|
| 14250 ± 4300 umol/L | SD media with 2% raffinose | 37 oC | BY4741 | PMID: 19174513 | | Conversion Details Here |
|
|---|
| Extracellular Concentrations | Not Available |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_2) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_3) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0gb9-5270921800-f4e2b38856bc1e6f4706 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0fvs-7391522200-c7937fce4dd0c2ae2a96 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a6s-9153231000-f6cd56307ab34ea34152 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udr-0190402200-1ed8e688ab88aec28e41 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0ug0-5291201000-59b06416b6afbd41045e | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9020100000-93f22c469a98f24b9da2 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014i-0000000900-59cc0160e38b2be8fd97 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0uxr-0191300900-676ca685bf43efd3f429 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0uxr-0391300900-978901884f71f0e0a53c | JSpectraViewer |
|
|---|
| References |
|---|
| References: | - Ejsing, C. S., Sampaio, J. L., Surendranath, V., Duchoslav, E., Ekroos, K., Klemm, R. W., Simons, K., Shevchenko, A. (2009). "Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry." Proc Natl Acad Sci U S A 106:2136-2141.19174513
|
|---|
| Synthesis Reference: | Not Available |
|---|
| External Links: | | Resource | Link |
|---|
| CHEBI ID | Not Available | | HMDB ID | HMDB10586 | | Pubchem Compound ID | 119120 | | Kegg ID | Not Available | | ChemSpider ID | 24768085 | | FOODB ID | FDB027736 | | Wikipedia ID | Not Available | | BioCyc ID | Not Available |
|
|---|
