| Identification |
|---|
| YMDB ID | YMDB00904 |
|---|
| Name | Pectin |
|---|
| Species | Saccharomyces cerevisiae |
|---|
| Strain | Baker's yeast |
|---|
| Description | Pectin, also known as galacturonate or D-lyxose, belongs to the class of organic compounds known as glucuronic acid derivatives. Glucuronic acid derivatives are compounds containing a glucuronic acid moiety (or a derivative), which consists of a glucose moiety with the C6 carbon oxidized to a carboxylic acid. Pectin is an extremely weak basic (essentially neutral) compound (based on its pKa). Pectin exists in all living species, ranging from bacteria to humans. |
|---|
| Structure | |
|---|
| Synonyms | - (+)-Xylose
- 2,3,4,5-Tetrahydroxypentanal
- D-Lyxose
- DL-Xylose
- L-Lyxose
- L(+)-Xylose
- Lyxose
- Pectin
- Pectin sugar
- Pectinose
- Pentose
- Trobicin
- Pectinic acid
- Calcium pectinate
- Methoxy pectin
- Methoxylpectin
- Methoxypectin
- Zinc pectinate
- Galacturonate
- b-D-Galacturonate
- b-D-Galacturonic acid
- beta-D-Galacturonate
- Β-D-galacturonate
- Β-D-galacturonic acid
- b-D-Galactopyranuronate
- b-D-Galactopyranuronic acid
- beta-D-Galactopyranuronate
- Β-D-galactopyranuronate
- Β-D-galactopyranuronic acid
|
|---|
| CAS number | 9000-69-5 |
|---|
| Weight | Average: 194.1394
Monoisotopic: 194.042652674 |
|---|
| InChI Key | AEMOLEFTQBMNLQ-DTEWXJGMSA-N |
|---|
| InChI | InChI=1S/C6H10O7/c7-1-2(8)4(5(10)11)13-6(12)3(1)9/h1-4,6-9,12H,(H,10,11)/t1-,2+,3+,4-,6+/m0/s1 |
|---|
| IUPAC Name | (2S,3R,4S,5R,6R)-3,4,5,6-tetrahydroxyoxane-2-carboxylic acid |
|---|
| Traditional IUPAC Name | β-D-galactopyranuronic acid |
|---|
| Chemical Formula | C6H10O7 |
|---|
| SMILES | [H]OC(=O)[C@@]1([H])O[C@@]([H])(O[H])[C@]([H])(O[H])[C@@]([H])(O[H])[C@@]1([H])O[H] |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as glucuronic acid derivatives. Glucuronic acid derivatives are compounds containing a glucuronic acid moiety (or a derivative), which consists of a glucose moiety with the C6 carbon oxidized to a carboxylic acid. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | Glucuronic acid derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Glucuronic acid or derivatives
- Beta-hydroxy acid
- Hydroxy acid
- Pyran
- Monosaccharide
- Oxane
- Hemiacetal
- Secondary alcohol
- Carboxylic acid derivative
- Carboxylic acid
- Oxacycle
- Organoheterocyclic compound
- Monocarboxylic acid or derivatives
- Polyol
- Hydrocarbon derivative
- Organic oxide
- Carbonyl group
- Alcohol
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Charge | 0 |
|---|
| Melting point | Not Available |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Water Solubility | Not Available | PhysProp | | LogP | Not Available | PhysProp |
|
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations | |
|---|
| Organoleptic Properties | Not Available |
|---|
| SMPDB Pathways | | Amino sugar and nucleotide sugar metabolism | PW002413 | 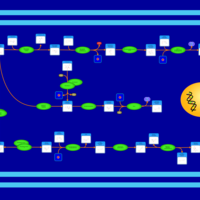 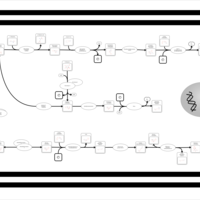 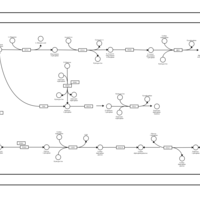 | | Starch and sucrose metabolism | PW002481 | 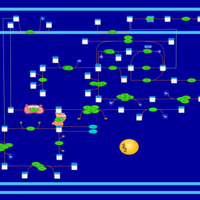 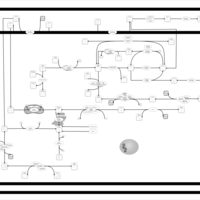 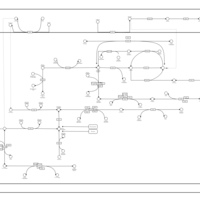 |
|
|---|
| KEGG Pathways | | Amino sugar and nucleotide sugar metabolism | ec00520 |  | | Pentose and glucuronate interconversions | ec00040 | 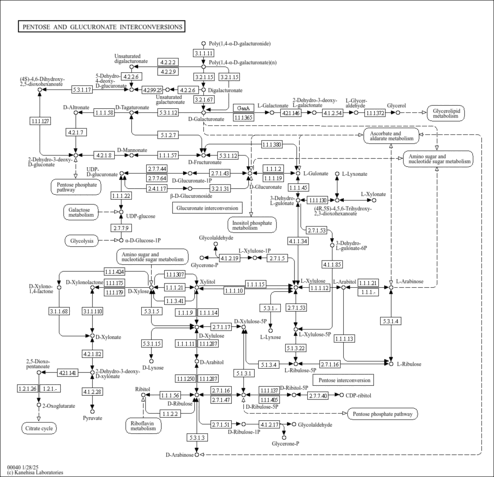 | | Starch and sucrose metabolism | ec00500 |  |
|
|---|
| SMPDB Reactions | Not Available |
|---|
| KEGG Reactions | |
|---|
| Concentrations |
|---|
| Intracellular Concentrations | Not Available |
|---|
| Extracellular Concentrations | Not Available |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0fvi-3900000000-623cfb10797e088f31a3 | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (5 TMS) - 70eV, Positive | splash10-000l-6242950000-7b610ed98440b47c2a32 | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | JSpectraViewer | | LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-00kr-6900000000-a4a0a279e2e16f432677 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-000i-9100000000-1c90dbe22f830f93ad0f | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-004i-9000000000-cefd20d23fb5136d2104 | JSpectraViewer | MoNA | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-002b-0900000000-142e51c19f7b152d18e6 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-056s-1900000000-80978d6e9cf43c2a99ae | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4r-9400000000-fe9d58fcfd8f4880243e | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0007-2900000000-eecfa94a9428872c9b56 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-000w-4900000000-c10f02b5bf83d62924f0 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-052f-9200000000-21b195d29543b18b8595 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0002-0900000000-2877a7e6d3ecd32cf172 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000b-7900000000-d748e59637b495442c9e | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03dl-9100000000-40ac961f184de816ed86 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-3900000000-64f3d0eb26f7bc8ca624 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a73-9300000000-329488f791f643b1c308 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4l-9000000000-474e82a5911096bc3b39 | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 2D NMR | [1H,1H] 2D NMR Spectrum | Not Available | JSpectraViewer | | 2D NMR | [1H,13C] 2D NMR Spectrum | Not Available | JSpectraViewer |
|
|---|
| References |
|---|
| References: | - Scheer, M., Grote, A., Chang, A., Schomburg, I., Munaretto, C., Rother, M., Sohngen, C., Stelzer, M., Thiele, J., Schomburg, D. (2011). "BRENDA, the enzyme information system in 2011." Nucleic Acids Res 39:D670-D676.21062828
- Herrgard, M. J., Swainston, N., Dobson, P., Dunn, W. B., Arga, K. Y., Arvas, M., Bluthgen, N., Borger, S., Costenoble, R., Heinemann, M., Hucka, M., Le Novere, N., Li, P., Liebermeister, W., Mo, M. L., Oliveira, A. P., Petranovic, D., Pettifer, S., Simeonidis, E., Smallbone, K., Spasic, I., Weichart, D., Brent, R., Broomhead, D. S., Westerhoff, H. V., Kirdar, B., Penttila, M., Klipp, E., Palsson, B. O., Sauer, U., Oliver, S. G., Mendes, P., Nielsen, J., Kell, D. B. (2008). "A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology." Nat Biotechnol 26:1155-1160.18846089
- Louw, C., Young, P. R., van Rensburg, P., Divol, B. (2010). "Regulation of endo-polygalacturonase activity in Saccharomyces cerevisiae." FEMS Yeast Res 10:44-57.19840115
|
|---|
| Synthesis Reference: | Gu, Qu-Ming; Nickol, Robert G.; Cheng, H. N. Enzyme-catalyzed modification of pectin. Polymer Preprints (American Chemical Society, Division of Polymer Chemistry) (2003), 44(2), 608-609. |
|---|
| External Links: | |
|---|



