| Identification |
|---|
| YMDB ID | YMDB00757 |
|---|
| Name | Glycerylphosphorylethanolamine |
|---|
| Species | Saccharomyces cerevisiae |
|---|
| Strain | Baker's yeast |
|---|
| Description | Glycerylphosphorylethanolamine, also known as GPEA, belongs to the class of organic compounds known as glycerophosphoethanolamines. These are glycerolipids characterized by an ethanolamine ester of glycerophosphoric acid. As is the case with diacylglycerols, glycerophosphoethanolamines can have many different combinations of fatty acids of varying lengths and saturation attached to the C-1 and C-2 atoms. Glycerylphosphorylethanolamine is a very strong basic compound (based on its pKa). |
|---|
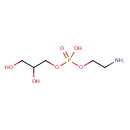
| Structure | |
|---|
| Synonyms | - 2-aminoethyl ester 1-Glycerophosphoric acid
- a-Glycerophosphorylethanolamine
- alpha-Glycerophosphorylethanolamine
- Glycerol 3-phosphoethanolamine
- Glycerol 3-phosphorylethanolamine
- Glycerophosphoethanolamine
- Glycerophosphorylethanolamine
- Glyceryl-3-phosphorylethanolamine
- Glycerylphosphorylethanolamine
- GPEA
- sn-glycero-3-Phosphoethanolamine
- sn-Glycerol-3-phosphoethanolamine
- Glycerol phosphorylethanolamine
- Α-glycerophosphorylethanolamine
|
|---|
| CAS number | Not Available |
|---|
| Weight | Average: 215.1415
Monoisotopic: 215.055873697 |
|---|
| InChI Key | JZNWSCPGTDBMEW-UHFFFAOYSA-N |
|---|
| InChI | InChI=1S/C5H14NO6P/c6-1-2-11-13(9,10)12-4-5(8)3-7/h5,7-8H,1-4,6H2,(H,9,10) |
|---|
| IUPAC Name | (2-aminoethoxy)(2,3-dihydroxypropoxy)phosphinic acid |
|---|
| Traditional IUPAC Name | glycerophosphorylethanolamine |
|---|
| Chemical Formula | C5H14NO6P |
|---|
| SMILES | [H]C(O)(CO)COP(O)(=O)OCCN |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as glycerophosphoethanolamines. These are glycerolipids characterized by an ethanolamine ester of glycerophosphoric acid. As is the case with diacylglycerols, glycerophosphoethanolamines can have many different combinations of fatty acids of varying lengths and saturation attached to the C-1 and C-2 atoms. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Glycerophospholipids |
|---|
| Sub Class | Glycerophosphoethanolamines |
|---|
| Direct Parent | Glycerophosphoethanolamines |
|---|
| Alternative Parents | |
|---|
| Substituents | - Sn-glycero-3-phosphoethanolamine
- Phosphoethanolamine
- Dialkyl phosphate
- Organic phosphoric acid derivative
- Phosphoric acid ester
- Alkyl phosphate
- Secondary alcohol
- 1,2-diol
- Primary amine
- Primary alcohol
- Organic oxygen compound
- Hydrocarbon derivative
- Organooxygen compound
- Organonitrogen compound
- Organic oxide
- Primary aliphatic amine
- Organic nitrogen compound
- Amine
- Organopnictogen compound
- Alcohol
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Charge | 0 |
|---|
| Melting point | Not Available |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Water Solubility | Not Available | PhysProp | | LogP | Not Available | PhysProp |
|
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations | |
|---|
| Organoleptic Properties | Not Available |
|---|
| SMPDB Pathways | Not Available |
|---|
| KEGG Pathways | Not Available |
|---|
| SMPDB Reactions | Not Available |
|---|
| KEGG Reactions | |
|---|
| Concentrations |
|---|
| Intracellular Concentrations | Not Available |
|---|
| Extracellular Concentrations | Not Available |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0gwf-5900000000-55796a83071cae1bff8b | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_5) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_6) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_7) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_2) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_3) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_4) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_1) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_2) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_3) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_4) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_5) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_6) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_7) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0006-9110000000-687d834f521d42c7e36c | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0006-9100000000-f3f7dcb5cdd10c48a24a | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-052f-9000000000-750b47541e6949c8a75f | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03di-2950000000-9e71c620f3a5db1afe86 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004l-9700000000-823f904dfffc25785d54 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-fdc87551301ed4f6a009 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0006-9710000000-e0cffde8c86999d55b58 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0007-9100000000-3885584fdb3534a0f84e | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9000000000-36cf29cb44eeb3400dfa | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0h00-9460000000-32cd47677e60fcdcf101 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9300000000-30cffc72754975634160 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-ca30e71e114ea6673088 | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer |
|
|---|
| References |
|---|
| References: | - Herrgard, M. J., Swainston, N., Dobson, P., Dunn, W. B., Arga, K. Y., Arvas, M., Bluthgen, N., Borger, S., Costenoble, R., Heinemann, M., Hucka, M., Le Novere, N., Li, P., Liebermeister, W., Mo, M. L., Oliveira, A. P., Petranovic, D., Pettifer, S., Simeonidis, E., Smallbone, K., Spasic, I., Weichart, D., Brent, R., Broomhead, D. S., Westerhoff, H. V., Kirdar, B., Penttila, M., Klipp, E., Palsson, B. O., Sauer, U., Oliver, S. G., Mendes, P., Nielsen, J., Kell, D. B. (2008). "A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology." Nat Biotechnol 26:1155-1160.18846089
- Angus, W. W., Lester, R. L. (1975). "The regulated catabolism of endogenous and exogenous phosphatidylinositol by Saccharomyces cerevisiae leading to extracellular glycerophosphorylinositol and inositol." J Biol Chem 250:22-30.166987
- Lee, K. S., Patton, J. L., Fido, M., Hines, L. K., Kohlwein, S. D., Paltauf, F., Henry, S. A., Levin, D. E. (1994). "The Saccharomyces cerevisiae PLB1 gene encodes a protein required for lysophospholipase and phospholipase B activity." J Biol Chem 269:19725-19730.8051052
|
|---|
| Synthesis Reference: | Not Available |
|---|
| External Links: | |
|---|