| Identification |
|---|
| YMDB ID | YMDB00727 |
|---|
| Name | (S)-2,3-epoxysqualene |
|---|
| Species | Saccharomyces cerevisiae |
|---|
| Strain | Baker's yeast |
|---|
| Description | (3S)-2,3-epoxy-2,3-dihydrosqualene, also known as (S)-squalene-2,3-epoxide, belongs to the class of organic compounds known as triterpenoids. These are terpene molecules containing six isoprene units (3S)-2,3-epoxy-2,3-dihydrosqualene is an extremely weak basic (essentially neutral) compound (based on its pKa). |
|---|
| Structure | |
|---|
| Synonyms | - (S)-2,3-Epoxysqualene
- (S)-Squalene-2,3-epoxide
- Squalene 2,3-epoxide
- Squalene 2,3-oxide
|
|---|
| CAS number | 9029-62-3 |
|---|
| Weight | Average: 426.7174
Monoisotopic: 426.386166222 |
|---|
| InChI Key | QYIMSPSDBYKPPY-RSKUXYSASA-N |
|---|
| InChI | InChI=1S/C30H50O/c1-24(2)14-11-17-27(5)20-12-18-25(3)15-9-10-16-26(4)19-13-21-28(6)22-23-29-30(7,8)31-29/h14-16,20-21,29H,9-13,17-19,22-23H2,1-8H3/b25-15+,26-16+,27-20+,28-21+/t29-/m0/s1 |
|---|
| IUPAC Name | (3S)-2,2-dimethyl-3-[(3Z,7E,11Z,15E)-3,7,12,16,20-pentamethylhenicosa-3,7,11,15,19-pentaen-1-yl]oxirane |
|---|
| Traditional IUPAC Name | (3S)-2,2-dimethyl-3-[(3Z,7E,11Z,15E)-3,7,12,16,20-pentamethylhenicosa-3,7,11,15,19-pentaen-1-yl]oxirane |
|---|
| Chemical Formula | C30H50O |
|---|
| SMILES | CC(C)=CCC\C(C)=C\CC\C(C)=C\CC\C=C(/C)CC\C=C(/C)CC[C@@H]1OC1(C)C |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as triterpenoids. These are terpene molecules containing six isoprene units. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Triterpenoids |
|---|
| Direct Parent | Triterpenoids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Triterpenoid
- Oxacycle
- Organoheterocyclic compound
- Ether
- Oxirane
- Dialkyl ether
- Organic oxygen compound
- Hydrocarbon derivative
- Organooxygen compound
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Charge | 0 |
|---|
| Melting point | Not Available |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Water Solubility | Not Available | PhysProp | | LogP | Not Available | PhysProp |
|
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations | - endoplasmic reticulum
- cytoplasm
|
|---|
| Organoleptic Properties | Not Available |
|---|
| SMPDB Pathways | | Cholesterol biosynthesis and metabolism CE(10:0) | PW002545 | 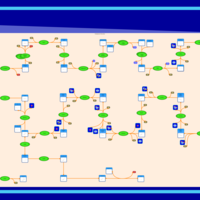 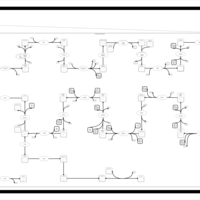 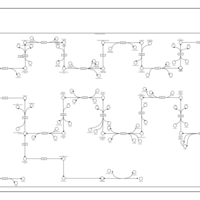 | | Cholesterol biosynthesis and metabolism CE(12:0) | PW002548 | 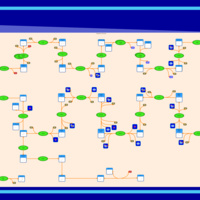 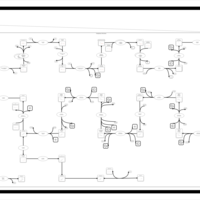 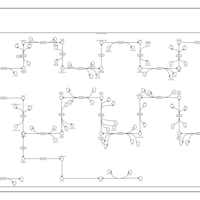 | | Cholesterol biosynthesis and metabolism CE(14:0) | PW002544 | 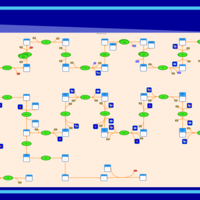 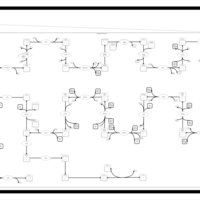 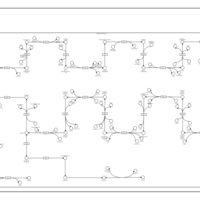 | | Cholesterol biosynthesis and metabolism CE(16:0) | PW002550 | 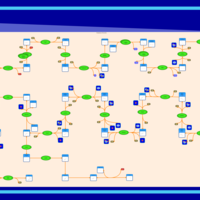 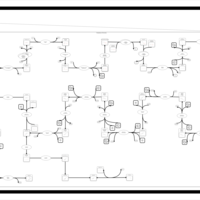 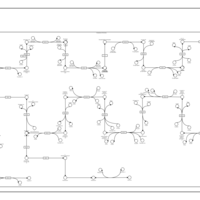 | | Cholesterol biosynthesis and metabolism CE(18:0) | PW002551 |    |
|
|---|
| KEGG Pathways | |
|---|
| SMPDB Reactions | |
|---|
| KEGG Reactions | |
|---|
| Concentrations |
|---|
| Intracellular Concentrations | Not Available |
|---|
| Extracellular Concentrations | Not Available |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-01rf-3696400000-dd04dae2363e5ff874a8 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-004i-0424900000-45b1f62177f69536d624 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-066r-6549200000-1fc45aff6d7829383ace | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-06dr-7492000000-8f3b1e8848877c4f3099 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-0000900000-180630471fad3d5ffc90 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-2000900000-727a64bd610baa87c554 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-9205300000-2ffaa263ecaf2ba728cf | JSpectraViewer |
|
|---|
| References |
|---|
| References: | - UniProt Consortium (2011). "Ongoing and future developments at the Universal Protein Resource." Nucleic Acids Res 39:D214-D219.21051339
- Scheer, M., Grote, A., Chang, A., Schomburg, I., Munaretto, C., Rother, M., Sohngen, C., Stelzer, M., Thiele, J., Schomburg, D. (2011). "BRENDA, the enzyme information system in 2011." Nucleic Acids Res 39:D670-D676.21062828
- Herrgard, M. J., Swainston, N., Dobson, P., Dunn, W. B., Arga, K. Y., Arvas, M., Bluthgen, N., Borger, S., Costenoble, R., Heinemann, M., Hucka, M., Le Novere, N., Li, P., Liebermeister, W., Mo, M. L., Oliveira, A. P., Petranovic, D., Pettifer, S., Simeonidis, E., Smallbone, K., Spasic, I., Weichart, D., Brent, R., Broomhead, D. S., Westerhoff, H. V., Kirdar, B., Penttila, M., Klipp, E., Palsson, B. O., Sauer, U., Oliver, S. G., Mendes, P., Nielsen, J., Kell, D. B. (2008). "A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology." Nat Biotechnol 26:1155-1160.18846089
- Lamb, D. C., Kelly, D. E., Manning, N. J., Kaderbhai, M. A., Kelly, S. L. (1999). "Biodiversity of the P450 catalytic cycle: yeast cytochrome b5/NADH cytochrome b5 reductase complex efficiently drives the entire sterol 14-demethylation (CYP51) reaction." FEBS Lett 462:283-288.10622712
|
|---|
| Synthesis Reference: | Not Available |
|---|
| External Links: | |
|---|

