| Identification |
|---|
| YMDB ID | YMDB00676 |
|---|
| Name | Caprylic acid |
|---|
| Species | Saccharomyces cerevisiae |
|---|
| Strain | Baker's yeast |
|---|
| Description | Caprylic acid, also known as octanoate or 8:0, belongs to the class of organic compounds known as medium-chain fatty acids. These are fatty acids with an aliphatic tail that contains between 4 and 12 carbon atoms. Caprylic acid is a very hydrophobic molecule, practically insoluble (in water), and relatively neutral. Caprylic acid is a potentially toxic compound. |
|---|
| Structure | |
|---|
| Synonyms | - 0ctanoic acid
- 1-Heptanecarboxylate
- 1-Heptanecarboxylic acid
- 1-octanoic acid
- 8-[(1R,2R)-3-Oxo-2-{(Z)-pent-2-enyl}cyclopentyl]octanoate
- Acide octanoique
- Acido octanoico
- Acidum octanocium
- C-8 Acid
- C8:0
- Caprylate
- Caprylic acid
- capryloate
- Caprylsaeure
- CH3-[CH2]6-COOH
- Enantic acid
- Heptane-1-carboxylic acid
- Kaprylsaeure
- Kyselina kaprylova
- n-Caprylate
- n-Caprylic acid
- n-Octanoate
- n-Octanoic acid
- n-Octoate
- n-Octoic acid
- n-Octylate
- n-Octylic acid
- octanoate
- Octanoic acid
- octanoic acid, ion(1-)
- Octansaeure
- Octic acid
- octoic acid
- Octylate
- octylic acid
- 8:0
- Acidum octanoicum
- OCTANOIC ACID (caprylIC ACID)
- OCTANOate (caprylate)
- Octoate
- Emery 657
- Kortacid 0899
- Lunac 8-95
- Lunac 8-98
- Neo-fat 8
- Neo-fat 8S
- Prifac 2901
- Caprylic acid, cadmium salt
- Caprylic acid, cesium salt
- Caprylic acid, manganese salt
- Caprylic acid, nickel(+2) salt
- Caprylic acid, zinc salt
- Caprylic acid, aluminum salt
- Caprylic acid, barium salt
- Caprylic acid, chromium(+2) salt
- Caprylic acid, lead(+2) salt
- Caprylic acid, potassium salt
- Caprylic acid, tin(+2) salt
- Sodium octanoate
- Caprylic acid, 14C-labeled
- Caprylic acid, lithium salt
- Caprylic acid, ruthenium(+3) salt
- Caprylic acid, sodium salt
- Caprylic acid, sodium salt, 11C-labeled
- Caprylic acid, tin salt
- Caprylic acid, zirconium salt
- Sodium caprylate
- Caprylic acid, ammonia salt
- Caprylic acid, calcium salt
- Caprylic acid, cobalt salt
- Caprylic acid, copper salt
- Caprylic acid, copper(+2) salt
- Caprylic acid, iridum(+3) salt
- Caprylic acid, iron(+3) salt
- Caprylic acid, lanthanum(+3) salt
- Caprylic acid, zirconium(+4) salt
- FA(8:0)
- Lithium octanoate
|
|---|
| CAS number | 124-07-2 |
|---|
| Weight | Average: 144.2114
Monoisotopic: 144.115029756 |
|---|
| InChI Key | WWZKQHOCKIZLMA-UHFFFAOYSA-N |
|---|
| InChI | InChI=1S/C8H16O2/c1-2-3-4-5-6-7-8(9)10/h2-7H2,1H3,(H,9,10) |
|---|
| IUPAC Name | octanoic acid |
|---|
| Traditional IUPAC Name | caprylic acid |
|---|
| Chemical Formula | C8H16O2 |
|---|
| SMILES | CCCCCCCC(O)=O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as medium-chain fatty acids. These are fatty acids with an aliphatic tail that contains between 4 and 12 carbon atoms. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Fatty Acyls |
|---|
| Sub Class | Fatty acids and conjugates |
|---|
| Direct Parent | Medium-chain fatty acids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Medium-chain fatty acid
- Straight chain fatty acid
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Carboxylic acid derivative
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Carbonyl group
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State | Liquid |
|---|
| Charge | 0 |
|---|
| Melting point | 16.5 °C |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Water Solubility | 0.789 mg/mL at 30 oC [YALKOWSKY,SH & DANNENFELSER,RM (1992)] | PhysProp | | LogP | 3.05 [HANSCH,C ET AL. (1995)] | PhysProp |
|
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations | - extracellular
- cell envelope
- mitochondrion
- endoplasmic reticulum
- lipid particle
- peroxisome
- cytoplasm
|
|---|
| Organoleptic Properties | |
|---|
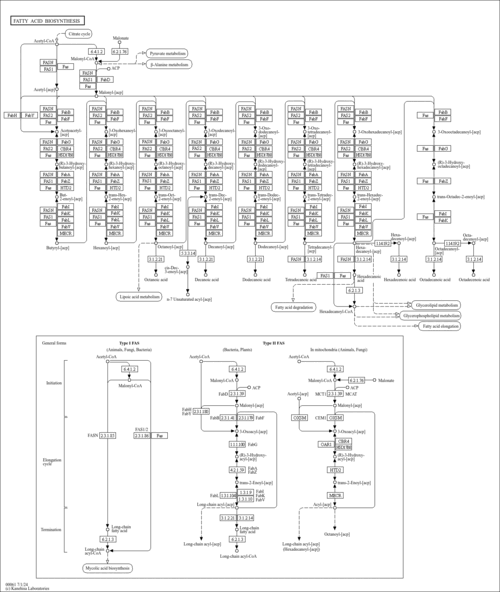
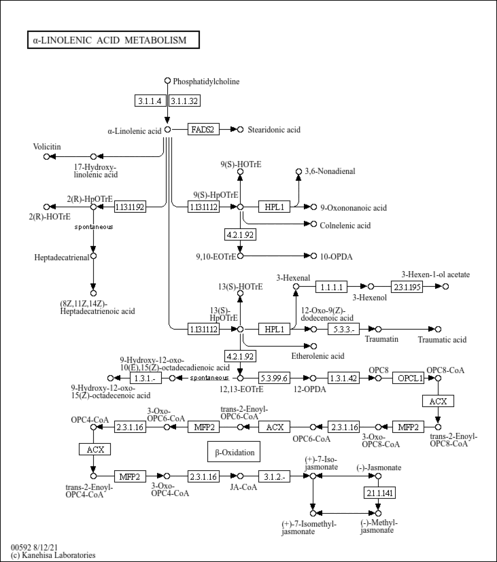
| SMPDB Pathways | |
|---|
| KEGG Pathways | |
|---|
| SMPDB Reactions | |
|---|
| KEGG Reactions | |
|---|
| Concentrations |
|---|
| Intracellular Concentrations | Not Available |
|---|
| Extracellular Concentrations | Not Available |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (Non-derivatized) | splash10-0159-0910000000-e609d5de69ede6fbcde0 | JSpectraViewer | MoNA | | GC-MS | GC-MS Spectrum - GC-MS (1 TMS) | splash10-0gb9-1920000000-1dc6ed976e003f99e23d | JSpectraViewer | MoNA | | GC-MS | GC-MS Spectrum - EI-B (Non-derivatized) | splash10-03kc-9000000000-74cfce03769b41313952 | JSpectraViewer | MoNA | | GC-MS | GC-MS Spectrum - EI-B (Non-derivatized) | splash10-0uyi-0931100000-7668e36e436054042b5c | JSpectraViewer | MoNA | | GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-0159-0910000000-e609d5de69ede6fbcde0 | JSpectraViewer | MoNA | | GC-MS | GC-MS Spectrum - GC-MS (Non-derivatized) | splash10-0gb9-1920000000-1dc6ed976e003f99e23d | JSpectraViewer | MoNA | | GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-014i-0910000000-9ecb4ccb46e89710f76a | JSpectraViewer | MoNA | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0596-9100000000-8854c56056d8d3405087 | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-062l-9100000000-67da39159980b9078fab | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | JSpectraViewer | | LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Negative (Annotated) | splash10-0006-0900000000-bb237630bedcee6145d1 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Negative (Annotated) | splash10-0006-2900000000-98284f50a271878c8481 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Negative (Annotated) | splash10-0006-5900000000-209fbbacd4996b9a770d | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - EI-B (HITACHI M-80B) , Positive | splash10-03kc-9000000000-74cfce03769b41313952 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Negative | splash10-0006-0900000000-e2c4dff10393086340f7 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Negative | splash10-0006-1900000000-bbde8564b4e5543a636a | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - DI-ESI-Q-Exactive Plus , Negative | splash10-0006-0900000000-d219a0c6559a4783fd81 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-0006-0900000000-e2c4dff10393086340f7 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-0006-1900000000-bbde8564b4e5543a636a | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-IT , negative | splash10-0006-0900000000-ca22ecd0b25fa371fc73 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-0gvp-9100000000-58c00a2024ba0bae4ff4 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-0002-9300000000-2505e6738cf5d5877654 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-0006-0900000000-1ab4cdf8f74c93312743 | JSpectraViewer | MoNA | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-002b-2900000000-ac7c80e50c93b8cc3af7 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0002-9400000000-d2694b5f89d8eaeb2ab1 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-052f-9000000000-f2b24072c91d8b0e4078 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-1900000000-ad62d44e72f4dd1760ab | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0007-7900000000-9c869b84815f21dffc7c | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4l-9000000000-b8a648d443695914e479 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0a6u-9100000000-f5c38fdad70b324e75ac | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a4i-9000000000-bee103956686b4840d1e | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4l-9000000000-e7cbd036c8e80018c825 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-0900000000-8f2b048da6331570976c | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0006-0900000000-cd31ca01a5fa6be86acb | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-052f-9100000000-682c215be5abe1fe87ed | JSpectraViewer | | MS | Mass Spectrum (Electron Ionization) | splash10-03kc-9000000000-8c7db9a9d75b2cc1bd1c | JSpectraViewer | MoNA | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 2D NMR | [1H,13C] 2D NMR Spectrum | Not Available | JSpectraViewer |
|
|---|
| References |
|---|
| References: | - Herrgard, M. J., Swainston, N., Dobson, P., Dunn, W. B., Arga, K. Y., Arvas, M., Bluthgen, N., Borger, S., Costenoble, R., Heinemann, M., Hucka, M., Le Novere, N., Li, P., Liebermeister, W., Mo, M. L., Oliveira, A. P., Petranovic, D., Pettifer, S., Simeonidis, E., Smallbone, K., Spasic, I., Weichart, D., Brent, R., Broomhead, D. S., Westerhoff, H. V., Kirdar, B., Penttila, M., Klipp, E., Palsson, B. O., Sauer, U., Oliver, S. G., Mendes, P., Nielsen, J., Kell, D. B. (2008). "A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology." Nat Biotechnol 26:1155-1160.18846089
- Furukawa, K., Yamada, T., Mizoguchi, H., Hara, S. (2003). "Increased ethyl caproate production by inositol limitation in Saccharomyces cerevisiae." J Biosci Bioeng 95:448-454.16233438
- Antonelli, A., Castellari, L., Zambonelli, C., Carnacini, A. (1999). "Yeast influence on volatile composition of wines." J Agric Food Chem 47:1139-1144.10552428
- Tsakiris, A., Koutinas, A. A., Psarianos, C., Kourkoutas, Y., Bekatorou, A. (2010). "A new process for wine production by penetration of yeast in uncrushed frozen grapes." Appl Biochem Biotechnol 162:1109-1121.20151225
- Patel, S., Shibamoto, T. (2002). "Effect of different strains of Saccharomyces cerevisiae on production of volatiles in Napa Gamay wine and Petite Sirah wine." J Agric Food Chem 50:5649-5653.12236692
- Kogan, G. L., Filipp, D., Arman, I. P., Leibovich, B. A., Beliaeva, E. S. (1991). "[Cloning of segments of the Drosophila melanogaster genome using artificial chromosomes of the yeast Saccharomyces cerevisiae]." Genetika 27:1316-1323.1761208
|
|---|
| Synthesis Reference: | Caprylic acid. Jpn. Kokai Tokkyo Koho (1983), 4 pp. |
|---|
| External Links: | |
|---|