| Identification |
|---|
| YMDB ID | YMDB00602 |
|---|
| Name | Biotinyl-5'-AMP |
|---|
| Species | Saccharomyces cerevisiae |
|---|
| Strain | Baker's yeast |
|---|
| Description | Biotinyl-5'-AMP belongs to the class of organic compounds known as 5'-acylphosphoadenosines. These are ribonucleoside derivatives containing an adenoside moiety, where the phosphate group is acylated. Biotinyl-5'-AMP is a strong basic compound (based on its pKa). Biotinyl-5'-AMP may be a unique S. cerevisiae (yeast) metabolite. |
|---|
| Structure | |
|---|
| Synonyms | - (+)-Biotinyl 5'-adenylate
- 5'-Adenylic acid anhydride with biotin
- 5'-Adenylic acid monoanhydride with biotin
- B-AMP
- beta-AMP
- bio-5-AMP
- Biotin anhydride with 5'-adenylic acid
- Biotinoyl 5'-adenylate
- Biotinyl 5'-AMP
- Biotinyl-5'-AMP
|
|---|
| CAS number | 4130-20-5 |
|---|
| Weight | Average: 573.517
Monoisotopic: 573.140682731 |
|---|
| InChI Key | UTQCSTJVMLODHM-RHCAYAJFSA-N |
|---|
| InChI | InChI=1S/C20H28N7O9PS/c21-17-14-18(23-7-22-17)27(8-24-14)19-16(30)15(29)10(35-19)5-34-37(32,33)36-12(28)4-2-1-3-11-13-9(6-38-11)25-20(31)26-13/h7-11,13,15-16,19,29-30H,1-6H2,(H,32,33)(H2,21,22,23)(H2,25,26,31)/t9-,10+,11-,13-,15+,16+,19+/m0/s1 |
|---|
| IUPAC Name | ({5-[(3aS,4S,6aR)-2-oxo-hexahydro-1H-thieno[3,4-d]imidazolidin-4-yl]pentanoyl}oxy)({[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy})phosphinic acid |
|---|
| Traditional IUPAC Name | β-amp |
|---|
| Chemical Formula | C20H28N7O9PS |
|---|
| SMILES | [H][C@]12CS[C@@H](CCCCC(=O)OP(O)(=O)OC[C@H]3O[C@H]([C@H](O)[C@@H]3O)N3C=NC4=C3N=CN=C4N)[C@@]1([H])NC(=O)N2 |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as 5'-acylphosphoadenosines. These are ribonucleoside derivatives containing an adenoside moiety, where the phosphate group is acylated. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Nucleosides, nucleotides, and analogues |
|---|
| Class | Purine nucleotides |
|---|
| Sub Class | Purine ribonucleotides |
|---|
| Direct Parent | 5'-acylphosphoadenosines |
|---|
| Alternative Parents | |
|---|
| Substituents | - 5'-acylphosphoadenosine
- Biotin_derivative
- Biotin
- Pentose phosphate
- Pentose-5-phosphate
- Glycosyl compound
- N-glycosyl compound
- 6-aminopurine
- Monosaccharide phosphate
- Pentose monosaccharide
- Imidazopyrimidine
- Purine
- Thienoimidazolidine
- Acyl phosphate
- Aminopyrimidine
- Monoalkyl phosphate
- Imidazolidinone
- Imidolactam
- Monosaccharide
- N-substituted imidazole
- Organic phosphoric acid derivative
- Phosphoric acid ester
- Alkyl phosphate
- Pyrimidine
- Thiophene
- Thiolane
- Tetrahydrofuran
- Azole
- Imidazole
- Imidazolidine
- Heteroaromatic compound
- Amino acid or derivatives
- Carbonic acid derivative
- Urea
- Secondary alcohol
- 1,2-diol
- Carboxylic acid salt
- Dialkylthioether
- Oxacycle
- Azacycle
- Monocarboxylic acid or derivatives
- Carboxylic acid derivative
- Thioether
- Organoheterocyclic compound
- Primary amine
- Hydrocarbon derivative
- Carbonyl group
- Organic oxygen compound
- Organic nitrogen compound
- Organic salt
- Organonitrogen compound
- Organooxygen compound
- Organopnictogen compound
- Organic oxide
- Alcohol
- Amine
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Charge | 0 |
|---|
| Melting point | Not Available |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Water Solubility | Not Available | PhysProp | | LogP | Not Available | PhysProp |
|
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations | |
|---|
| Organoleptic Properties | Not Available |
|---|
| SMPDB Pathways | Not Available |
|---|
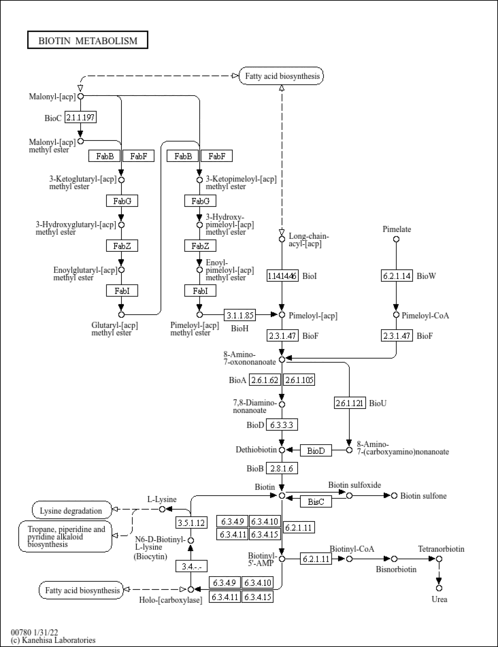
| KEGG Pathways | |
|---|
| SMPDB Reactions | Not Available |
|---|
| KEGG Reactions | |
|---|
| Concentrations |
|---|
| Intracellular Concentrations | Not Available |
|---|
| Extracellular Concentrations | Not Available |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-0932030000-9fc6a8392ffae33e1669 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-0910000000-2446591c42df4f29b9dc | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-2900000000-4520d832999df4668da5 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00c0-2319030000-2145defc8444c115071f | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-001i-6902000000-b1fc9db8b42178ed1000 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-003r-9500000000-184eb594d205789edea6 | JSpectraViewer |
|
|---|
| References |
|---|
| References: | - Herrgard, M. J., Swainston, N., Dobson, P., Dunn, W. B., Arga, K. Y., Arvas, M., Bluthgen, N., Borger, S., Costenoble, R., Heinemann, M., Hucka, M., Le Novere, N., Li, P., Liebermeister, W., Mo, M. L., Oliveira, A. P., Petranovic, D., Pettifer, S., Simeonidis, E., Smallbone, K., Spasic, I., Weichart, D., Brent, R., Broomhead, D. S., Westerhoff, H. V., Kirdar, B., Penttila, M., Klipp, E., Palsson, B. O., Sauer, U., Oliver, S. G., Mendes, P., Nielsen, J., Kell, D. B. (2008). "A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology." Nat Biotechnol 26:1155-1160.18846089
- Hoja, U., Wellein, C., Greiner, E., Schweizer, E. (1998). "Pleiotropic phenotype of acetyl-CoA-carboxylase-defective yeast cells--viability of a BPL1-amber mutation depending on its readthrough by normal tRNA(Gln)(CAG)." Eur J Biochem 254:520-526.9688262
|
|---|
| Synthesis Reference: | Christner, James E.; Coon, Minor J. Synthesis of (+)-biotinyl 5'-adenylate. Methods Enzymol. (1970), 18(Pt. A), 386-90. |
|---|
| External Links: | | Resource | Link |
|---|
| CHEBI ID | 3110 | | HMDB ID | HMDB04220 | | Pubchem Compound ID | 440839 | | Kegg ID | C05921 | | ChemSpider ID | Not Available | | FOODB ID | Not Available | | Wikipedia ID | Not Available | | BioCyc ID | Not Available |
|
|---|