palmitoyl-CoA (YMDB00527)
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| YMDB ID | YMDB00527 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name | palmitoyl-CoA | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Species | Saccharomyces cerevisiae | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Strain | Baker's yeast | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | hexadecanoyl-CoA belongs to the class of organic compounds known as hexoses. These are monosaccharides in which the sugar unit is a is a six-carbon containing moeity. Thus, hexadecanoyl-CoA is considered to be a fatty ester lipid molecule. hexadecanoyl-CoA is a very hydrophobic molecule, practically insoluble (in water), and relatively neutral. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number | 1763-10-6 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Weight | Average: 1005.943 Monoisotopic: 1005.344873947 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key | MNBKLUUYKPBKDU-BBECNAHFSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI | InChI=1S/C37H66N7O17P3S/c1-4-5-6-7-8-9-10-11-12-13-14-15-16-17-28(46)65-21-20-39-27(45)18-19-40-35(49)32(48)37(2,3)23-58-64(55,56)61-63(53,54)57-22-26-31(60-62(50,51)52)30(47)36(59-26)44-25-43-29-33(38)41-24-42-34(29)44/h24-26,30-32,36,47-48H,4-23H2,1-3H3,(H,39,45)(H,40,49)(H,53,54)(H,55,56)(H2,38,41,42)(H2,50,51,52)/t26-,30-,31-,32+,36-/m1/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name | {[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-2-({[({[(3R)-3-[(2-{[2-(hexadecanoylsulfanyl)ethyl]carbamoyl}ethyl)carbamoyl]-3-hydroxy-2,2-dimethylpropoxy](hydroxy)phosphoryl}oxy)(hydroxy)phosphoryl]oxy}methyl)-4-hydroxyoxolan-3-yl]oxy}phosphonic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name | palmitoyl-coa | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula | C37H66N7O17P3S | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES | CCCCCCCCCCCCCCCC(=O)SCCNC(=O)CCNC(=O)[C@H](O)C(C)(C)COP(O)(=O)OP(O)(=O)OC[C@H]1O[C@H]([C@H](O)[C@@H]1OP(O)(O)=O)N1C=NC2=C1N=CN=C2N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as hexoses. These are monosaccharides in which the sugar unit is a is a six-carbon containing moeity. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organic oxygen compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Organooxygen compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Carbohydrates and carbohydrate conjugates | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Hexoses | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic heteromonocyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge | 0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Organoleptic Properties | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMPDB Pathways |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
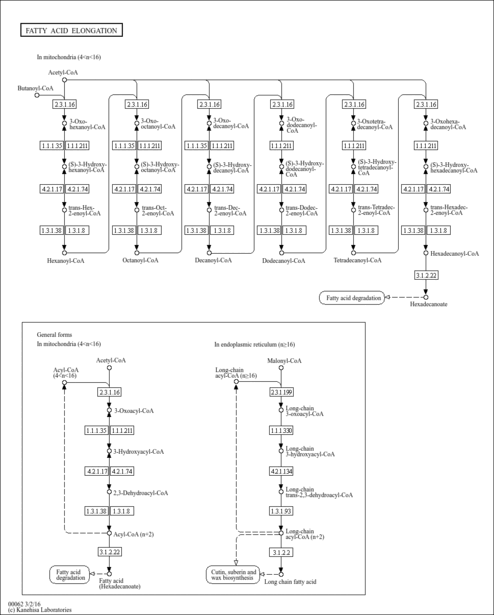
| KEGG Pathways |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMPDB Reactions | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG Reactions | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Concentrations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Intracellular Concentrations | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Extracellular Concentrations | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in catalytic activity
- Specific function:
- Esterification, concomitant with transport, of exogenous long-chain fatty acids into metabolically active CoA thioesters for subsequent degradation or incorporation into phospholipids. Contributes, with FAA1, to the activation of imported myristate
- Gene Name:
- FAA4
- Uniprot ID:
- P47912
- Molecular weight:
- 77266.5
Reactions
| ATP + a long-chain carboxylic acid + CoA → AMP + diphosphate + an acyl-CoA. |
- General function:
- Involved in transferase activity, transferring acyl groups
- Specific function:
- Probable acetyltransferase, which acetylates the inositol ring of phosphatidylinositol during biosynthesis of GPI- anchor. Acetylation during GPI-anchor biosynthesis is not essential for the subsequent mannosylation and is usually removed soon after the attachment of GPIs to proteins
- Gene Name:
- GWT1
- Uniprot ID:
- P47026
- Molecular weight:
- 55465.60156
Reactions
- General function:
- Involved in transferase activity, transferring nitrogenous groups
- Specific function:
- Component of serine palmitoyltransferase (SPT), which catalyzes the committed step in the synthesis of sphingolipids, the condensation of serine with palmitoyl CoA to form the long chain base 3-ketosphinganine
- Gene Name:
- LCB1
- Uniprot ID:
- P25045
- Molecular weight:
- 62206.60156
Reactions
| Palmitoyl-CoA + L-serine → CoA + 3-dehydro-D-sphinganine + CO(2). |
- General function:
- Involved in acyl-CoA dehydrogenase activity
- Specific function:
- Acyl-CoA + O(2) = trans-2,3-dehydroacyl-CoA + H(2)O(2)
- Gene Name:
- POX1
- Uniprot ID:
- P13711
- Molecular weight:
- 84041.39844
Reactions
| Acyl-CoA + O(2) → trans-2,3-dehydroacyl-CoA + H(2)O(2). |
- General function:
- Involved in catalytic activity
- Specific function:
- Esterification, concomitant with transport, of exogenous long-chain fatty acids into metabolically active CoA thioesters for subsequent degradation or incorporation into phospholipids. It may supplement intracellular myristoyl-CoA pools from exogenous myristate. Preferentially acts on C12:0-C16:0 fatty acids with myristic and pentadecanic acid (C15:0) having the highest activities
- Gene Name:
- FAA1
- Uniprot ID:
- P30624
- Molecular weight:
- 77865.79688
Reactions
| ATP + a long-chain carboxylic acid + CoA → AMP + diphosphate + an acyl-CoA. |
- General function:
- Involved in catalytic activity
- Specific function:
- Esterification, concomitant with transport, of endogenous long-chain fatty acids into metabolically active CoA thioesters for subsequent degradation or incorporation into phospholipids. This enzyme acts preferentially on C16 and C18 fatty acids with a cis-double bond at C-9-C-10
- Gene Name:
- FAA3
- Uniprot ID:
- P39002
- Molecular weight:
- 77946.0
Reactions
| ATP + a long-chain carboxylic acid + CoA → AMP + diphosphate + an acyl-CoA. |
- General function:
- Involved in catalytic activity
- Specific function:
- Esterification, concomitant with transport, of endogenous long-chain fatty acids into metabolically active CoA thioesters for subsequent degradation or incorporation into phospholipids. Preferentially acts on C9:0-C13:0 fatty acids although C7:0-C17:0 fatty acids are tolerated
- Gene Name:
- FAA2
- Uniprot ID:
- P39518
- Molecular weight:
- 83437.10156
Reactions
| ATP + a long-chain carboxylic acid + CoA → AMP + diphosphate + an acyl-CoA. |
- General function:
- Involved in transferase activity, transferring acyl groups other than amino-acyl groups
- Specific function:
- Acyl-CoA + acetyl-CoA = CoA + 3-oxoacyl-CoA
- Gene Name:
- POT1
- Uniprot ID:
- P27796
- Molecular weight:
- 44729.89844
Reactions
| Acyl-CoA + acetyl-CoA → CoA + 3-oxoacyl-CoA. |
- General function:
- Involved in transferase activity
- Specific function:
- Fatty acid synthetase catalyzes the formation of long- chain fatty acids from acetyl-CoA, malonyl-CoA and NADPH. The beta subunit contains domains for:[acyl-carrier-protein] acetyltransferase and malonyltransferase, S-acyl fatty acid synthase thioesterase, enoyl-[acyl-carrier-protein] reductase, and 3-hydroxypalmitoyl-[acyl-carrier-protein] dehydratase
- Gene Name:
- FAS1
- Uniprot ID:
- P07149
- Molecular weight:
- 228689.0
Reactions
| Acetyl-CoA + n malonyl-CoA + 2n NADH + 2n NADPH → long-chain-acyl-CoA + n CoA + n CO(2) + 2n NAD(+) + 2n NADP(+). |
| Acetyl-CoA + [acyl-carrier-protein] → CoA + acetyl-[acyl-carrier-protein]. |
| Malonyl-CoA + [acyl-carrier-protein] → CoA + malonyl-[acyl-carrier-protein]. |
| (3R)-3-hydroxypalmitoyl-[acyl-carrier-protein] → hexadec-2-enoyl-[acyl-carrier-protein] + H(2)O. |
| Acyl-[acyl-carrier-protein] + NAD(+) → trans-2,3-dehydroacyl-[acyl-carrier-protein] + NADH. |
| Oleoyl-[acyl-carrier-protein] + H(2)O → [acyl-carrier-protein] + oleate. |
- General function:
- Involved in transferase activity
- Specific function:
- Catalytic subunit of serine palmitoyltransferase (SPT), which catalyzes the committed step in the synthesis of sphingolipids, the condensation of serine with palmitoyl CoA to form the long chain base 3-ketosphinganine
- Gene Name:
- LCB2
- Uniprot ID:
- P40970
- Molecular weight:
- 63110.19922
Reactions
| Palmitoyl-CoA + L-serine → CoA + 3-dehydro-D-sphinganine + CO(2). |
- General function:
- Involved in acyltransferase activity
- Specific function:
- G-3-P/dihydroxyacetone phosphate dual substrate-specific sn-1 acyltransferase
- Gene Name:
- GPT1
- Uniprot ID:
- P32784
- Molecular weight:
- 85693.5
Reactions
| Acyl-CoA + sn-glycerol 3-phosphate → CoA + 1-acyl-sn-glycerol 3-phosphate. |
| Acyl-CoA + glycerone phosphate → CoA + acylglycerone phosphate. |
- General function:
- Involved in acyl-CoA thioesterase activity
- Specific function:
- Acyl-CoA thioesterases are a group of enzymes that catalyze the hydrolysis of acyl-CoAs to the free fatty acid and coenzyme A (CoASH), providing the potential to regulate intracellular levels of acyl-CoAs, free fatty acids and CoASH
- Gene Name:
- TES1
- Uniprot ID:
- P41903
- Molecular weight:
- 40259.39844
Reactions
| Palmitoyl-CoA + H(2)O → CoA + palmitate. |
- General function:
- Involved in acyltransferase activity
- Specific function:
- G-3-P/dihydroxyacetone phosphate dual substrate-specific sn-1 acyltransferase
- Gene Name:
- GPT2
- Uniprot ID:
- P36148
- Molecular weight:
- 83644.0
Reactions
| Acyl-CoA + sn-glycerol 3-phosphate → CoA + 1-acyl-sn-glycerol 3-phosphate. |
| Acyl-CoA + glycerone phosphate → CoA + acylglycerone phosphate. |
- General function:
- Involved in stearoyl-CoA 9-desaturase activity
- Specific function:
- Utilizes O(2) and electrons from the reduced cytochrome b(5) domain to catalyze the insertion of a double bond into a spectrum of fatty acyl-CoA substrates (Probable)
- Gene Name:
- OLE1
- Uniprot ID:
- P21147
- Molecular weight:
- 58402.60156
Reactions
| Stearoyl-CoA + 2 ferrocytochrome b5 + O(2) + 2 H(+) → oleoyl-CoA + 2 ferricytochrome b5 + 2 H(2)O. |
Transporters
- General function:
- Involved in transport
- Specific function:
- Involved in the import of activated long-chain fatty acids from the cytosol to the peroxisomal matrix
- Gene Name:
- PXA2
- Uniprot ID:
- P34230
- Molecular weight:
- 97125.29688