| Identification |
|---|
| YMDB ID | YMDB00357 |
|---|
| Name | Dephospho-CoA |
|---|
| Species | Saccharomyces cerevisiae |
|---|
| Strain | Baker's yeast |
|---|
| Description | Dephospho-CoA, also known as 3'-dephospho-CoA, belongs to the class of organic compounds known as purine ribonucleoside diphosphates. These are purine ribobucleotides with diphosphate group linked to the ribose moiety. Thus, dephospho-CoA is considered to be a fatty ester lipid molecule. Dephospho-CoA is a very hydrophobic molecule, practically insoluble (in water), and relatively neutral. Dephospho-CoA exists in all living species, ranging from bacteria to humans. Within yeast, dephospho-CoA participates in a number of enzymatic reactions. In particular, dephospho-CoA can be biosynthesized from 4'-phosphopantetheine; which is mediated by the enzyme phosphopantetheine adenylyltransferase. In addition, dephospho-CoA can be converted into coenzyme A; which is mediated by the enzyme dephospho-CoA kinase. In yeast, dephospho-CoA is involved in the metabolic pathway called pantothenate and coa biosynthesis pathway. |
|---|
| Structure | |
|---|
| Synonyms | - 3'-dephospho-CoA
- 3'-dephospho-Coenzyme A
- 3'-O-dephosphono-CoA
- 3'-O-dephosphono-Coenzyme A
- Dephospho-CoA
- Dephospho-Coenzyme A
- dephosphocoenzyme a
- DpCoA
- Dephospho CoA
- 3'-Desphospho-coenzyme A
- 2'-(5''-Triphosphoribosyl)-3'-dephospho-CoA
- Desphospho-CoA
|
|---|
| CAS number | 3633-59-8 |
|---|
| Weight | Average: 687.554
Monoisotopic: 687.148877955 |
|---|
| InChI Key | KDTSHFARGAKYJN-DRCCLKDXSA-N |
|---|
| InChI | InChI=1S/C21H35N7O13P2S/c1-21(2,16(32)19(33)24-4-3-12(29)23-5-6-44)8-39-43(36,37)41-42(34,35)38-7-11-14(30)15(31)20(40-11)28-10-27-13-17(22)25-9-26-18(13)28/h9-11,14-16,20,30-32,44H,3-8H2,1-2H3,(H,23,29)(H,24,33)(H,34,35)(H,36,37)(H2,22,25,26)/t11-,14-,15-,16?,20-/m1/s1 |
|---|
| IUPAC Name | [({[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy][3-hydroxy-2,2-dimethyl-3-({2-[(2-sulfanylethyl)carbamoyl]ethyl}carbamoyl)propoxy]phosphinic acid |
|---|
| Traditional IUPAC Name | {[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy(hydroxy)phosphoryl}oxy(3-hydroxy-2,2-dimethyl-3-({2-[(2-sulfanylethyl)carbamoyl]ethyl}carbamoyl)propoxy)phosphinic acid |
|---|
| Chemical Formula | C21H35N7O13P2S |
|---|
| SMILES | [H]OC([H])(C(=O)N([H])C([H])([H])C([H])([H])C(=O)N([H])C([H])([H])C([H])([H])S[H])C(C([H])([H])[H])(C([H])([H])[H])C([H])([H])OP(=O)(O[H])OP(=O)(O[H])OC([H])([H])[C@@]1([H])O[C@@]([H])(N2C([H])=NC3=C2N=C([H])N=C3N([H])[H])[C@]([H])(O[H])[C@]1([H])O[H] |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as purine ribonucleoside diphosphates. These are purine ribobucleotides with diphosphate group linked to the ribose moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Nucleosides, nucleotides, and analogues |
|---|
| Class | Purine nucleotides |
|---|
| Sub Class | Purine ribonucleotides |
|---|
| Direct Parent | Purine ribonucleoside diphosphates |
|---|
| Alternative Parents | |
|---|
| Substituents | - Purine ribonucleoside diphosphate
- Purine ribonucleoside monophosphate
- Pentose phosphate
- Pentose-5-phosphate
- Beta amino acid or derivatives
- Glycosyl compound
- N-glycosyl compound
- 6-aminopurine
- Pentose monosaccharide
- Monosaccharide phosphate
- Organic pyrophosphate
- Imidazopyrimidine
- Purine
- Aminopyrimidine
- Monoalkyl phosphate
- N-acyl-amine
- N-substituted imidazole
- Organic phosphoric acid derivative
- Pyrimidine
- Fatty amide
- Imidolactam
- Monosaccharide
- Fatty acyl
- Phosphoric acid ester
- Alkyl phosphate
- Azole
- Heteroaromatic compound
- Imidazole
- Tetrahydrofuran
- Secondary alcohol
- Secondary carboxylic acid amide
- Amino acid or derivatives
- Carboxamide group
- Organoheterocyclic compound
- Carboxylic acid derivative
- Alkylthiol
- Oxacycle
- Azacycle
- Organic oxide
- Organooxygen compound
- Organosulfur compound
- Carbonyl group
- Organopnictogen compound
- Organic nitrogen compound
- Organic oxygen compound
- Hydrocarbon derivative
- Alcohol
- Organonitrogen compound
- Primary amine
- Amine
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Charge | 0 |
|---|
| Melting point | Not Available |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Water Solubility | Not Available | PhysProp | | LogP | Not Available | PhysProp |
|
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations | - mitochondrion
- lipid particle
- endoplasmic reticulum
- cytoplasm
|
|---|
| Organoleptic Properties | Not Available |
|---|
| SMPDB Pathways | |
|---|
| KEGG Pathways | | Pantothenate and CoA biosynthesis | ec00770 | 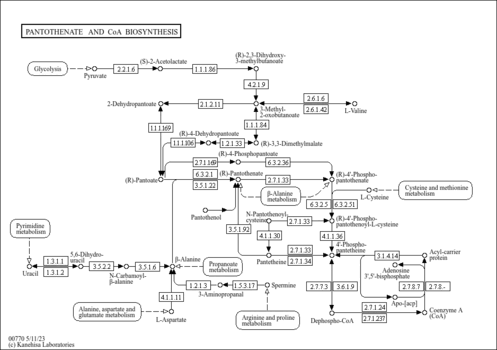 | | beta-Alanine metabolism | ec00410 |  |
|
|---|
| SMPDB Reactions | |
|---|
| KEGG Reactions | |
|---|
| Concentrations |
|---|
| Intracellular Concentrations | Not Available |
|---|
| Extracellular Concentrations | Not Available |
|---|
| Spectra |
|---|
| Spectra | |
|---|
| References |
|---|
| References: | - UniProt Consortium (2011). "Ongoing and future developments at the Universal Protein Resource." Nucleic Acids Res 39:D214-D219.21051339
|
|---|
| Synthesis Reference: | Not Available |
|---|
| External Links: | |
|---|


