| Identification |
|---|
| YMDB ID | YMDB00268 |
|---|
| Name | 4,4-Dimethylcholesta-8,14,24-trienol |
|---|
| Species | Saccharomyces cerevisiae |
|---|
| Strain | Baker's yeast |
|---|
| Description | 4,4-Dimethylcholesta-8,14,24-trienol, also known as FF-MAS or follicular fluid meiosis activating sterol, belongs to the class of organic compounds known as cholesterols and derivatives. Cholesterols and derivatives are compounds containing a 3-hydroxylated cholestane core. Thus, 4,4-dimethylcholesta-8,14,24-trienol is considered to be a sterol. Based on a literature review very few articles have been published on 4,4-Dimethylcholesta-8,14,24-trienol. |
|---|
| Structure | |
|---|
| Synonyms | - (3beta,5alpha)-4,4-dimethyl-Cholesta-8,14,24-trien-3-ol
- (3beta,5alpha)-4,4-Dimethylcholesta-8,14,24-trien-3-ol
- 4,4-Dimechol-8,14,24-trienol
- 4,4-dimethyl-5-alpha-cholesta-8,14,24-trien-3-beta-ol
- 4,4-dimethyl-5alpha-cholesta-8,14,24-trien-3-ol
- 4,4-Dimethyl-5alpha-cholesta-8,14,24-trien-3beta-ol
- 4,4-dimethyl-cholesta-8,14,24-trienol
- 4,4-Dimethylcholesta-8,14,24-trienol
- 4,4'-dimethyl cholesta-8,14,24-triene-3-beta-ol
- FF-MAS
- follicular fluid meiosis activating sterol
- 4,4-Dimethylcholesta-8(9),14,24-trien-3beta-ol
- 4,4-Dimethylcholesta-8(9),14,24-trien-3b-ol
- 4,4-Dimethylcholesta-8(9),14,24-trien-3β-ol
|
|---|
| CAS number | 64284-64-6 |
|---|
| Weight | Average: 410.6749
Monoisotopic: 410.354866094 |
|---|
| InChI Key | LFQXEZVYNCBVDO-PBJLWWPKSA-N |
|---|
| InChI | InChI=1S/C29H46O/c1-19(2)9-8-10-20(3)22-12-13-23-21-11-14-25-27(4,5)26(30)16-18-29(25,7)24(21)15-17-28(22,23)6/h9,13,20,22,25-26,30H,8,10-12,14-18H2,1-7H3/t20-,22-,25+,26+,28-,29-/m1/s1 |
|---|
| IUPAC Name | (2S,5S,7R,14R,15R)-2,6,6,15-tetramethyl-14-[(2R)-6-methylhept-5-en-2-yl]tetracyclo[8.7.0.0^{2,7}.0^{11,15}]heptadeca-1(10),11-dien-5-ol |
|---|
| Traditional IUPAC Name | (2S,5S,7R,14R,15R)-2,6,6,15-tetramethyl-14-[(2R)-6-methylhept-5-en-2-yl]tetracyclo[8.7.0.0^{2,7}.0^{11,15}]heptadeca-1(10),11-dien-5-ol |
|---|
| Chemical Formula | C29H46O |
|---|
| SMILES | [H]O[C@@]1([H])C([H])([H])C([H])([H])[C@@]2(C3=C(C4=C([H])C([H])([H])[C@]([H])([C@]([H])(C([H])([H])[H])C([H])([H])C([H])([H])C([H])=C(C([H])([H])[H])C([H])([H])[H])[C@@]4(C([H])([H])[H])C([H])([H])C3([H])[H])C([H])([H])C([H])([H])[C@@]2([H])C1(C([H])([H])[H])C([H])([H])[H])C([H])([H])[H] |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as cholesterols and derivatives. Cholesterols and derivatives are compounds containing a 3-hydroxylated cholestane core. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Steroids and steroid derivatives |
|---|
| Sub Class | Cholestane steroids |
|---|
| Direct Parent | Cholesterols and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Cholesterol-skeleton
- 3-beta-hydroxysteroid
- Hydroxysteroid
- 3-hydroxysteroid
- Cyclic alcohol
- Secondary alcohol
- Organic oxygen compound
- Hydrocarbon derivative
- Organooxygen compound
- Alcohol
- Aliphatic homopolycyclic compound
|
|---|
| Molecular Framework | Aliphatic homopolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Charge | 0 |
|---|
| Melting point | Not Available |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Water Solubility | Not Available | PhysProp | | LogP | Not Available | PhysProp |
|
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations | |
|---|
| Organoleptic Properties | Not Available |
|---|
| SMPDB Pathways | | Cholesterol biosynthesis and metabolism CE(10:0) | PW002545 | 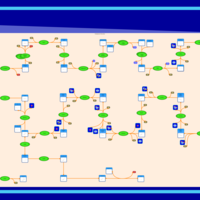 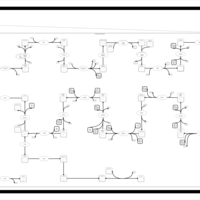 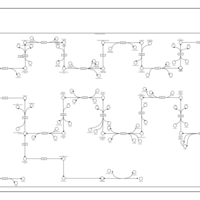 | | Cholesterol biosynthesis and metabolism CE(12:0) | PW002548 | 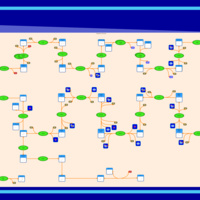 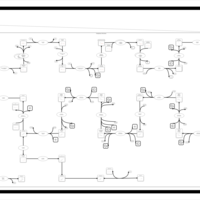 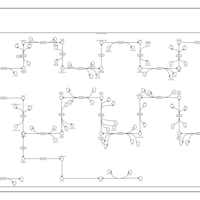 | | Cholesterol biosynthesis and metabolism CE(14:0) | PW002544 |  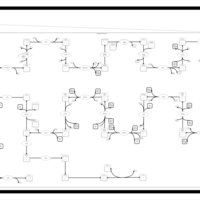 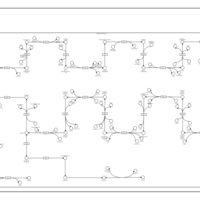 | | Cholesterol biosynthesis and metabolism CE(16:0) | PW002550 | 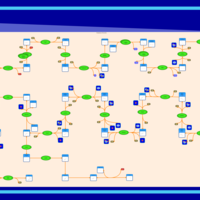 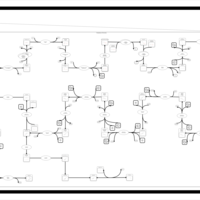 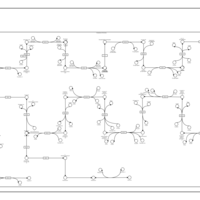 | | Cholesterol biosynthesis and metabolism CE(18:0) | PW002551 | 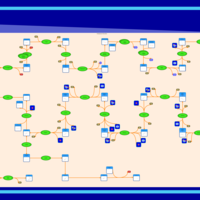 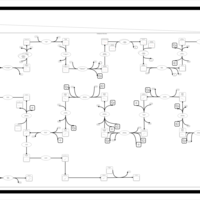 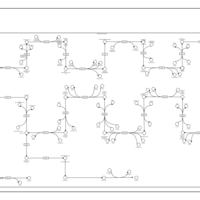 |
|
|---|
| KEGG Pathways | |
|---|
| SMPDB Reactions | |
|---|
| KEGG Reactions | | 4,4-Dimethylcholesta-8,14,24-trienol
+
NADPH
+
hydron
→
NADP
+
4,4-Dimethyl-5a-cholesta-8,24-dien-3-b-ol
| | oxygen
+
NADH
+
hydron
+
Lanosterin
→
water
+
Formic acid
+
NAD
+
4,4-Dimethylcholesta-8,14,24-trienol
| | oxygen
+
hydron
+
NADPH
+
Lanosterin
→
NADP
+
water
+
Formic acid
+
4,4-Dimethylcholesta-8,14,24-trienol
|
|
|---|
| Concentrations |
|---|
| Intracellular Concentrations | Not Available |
|---|
| Extracellular Concentrations | Not Available |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-000t-1009000000-416b93fbfcc925dc4766 | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-014i-4103900000-a8896cb66364cceda294 | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | JSpectraViewer | | LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 18V, positive | splash10-0006-1329000000-9b8610b57655e3a41ca1 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 22V, positive | splash10-0006-3958000000-39386b42d7d020c29bbb | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 25V, positive | splash10-05o3-4943000000-68305bc3d9c2e79f17bc | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 28V, positive | splash10-067j-4931000000-17ac98d22705a30ac479 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 32V, positive | splash10-0aos-4930000000-57d6f877cf0915f132b3 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 38V, positive | splash10-0aos-5910000000-64fc1bcaeb9b8921df29 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 48V, positive | splash10-0aou-5900000000-7ebf0af9fd57cfad6303 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 57V, positive | splash10-0ar3-4900000000-2533caeaeb9ebdebc4a1 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 70V, positive | splash10-054o-4900000000-bbdc94435c78cc8bc333 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 83V, positive | splash10-056u-4900000000-36892883de61aea8c544 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 102V, positive | splash10-00ou-4900000000-18c114c240680434a0d3 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - n/a 28V, positive | splash10-053r-0393000000-0c6dea5c7ec2bee79e1e | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - n/a 28V, positive | splash10-000l-0970000000-b2c5bf7eb779387cd981 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - n/a 28V, positive | splash10-006t-0930000000-e231f282c3639a162ab0 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - n/a 28V, positive | splash10-0aor-2900000000-85f247eedd1281442c90 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - n/a 28V, positive | splash10-006t-0950000000-15e1c7c9cb681913720e | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - n/a 28V, positive | splash10-0api-0900000000-eb024b4e1b55ee575bb2 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - n/a 28V, positive | splash10-0a4i-0900000000-6034a459e48936a045eb | JSpectraViewer | MoNA | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0002900000-3773f1780255ea800890 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4i-0005900000-43378c95eaacf13d3cd4 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004l-1009000000-1387c3c0fba5a00f56a6 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0000900000-84eae2c8ad8ccafd7741 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4i-0000900000-84eae2c8ad8ccafd7741 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-0013900000-62d1d4a75b4aa2cd0a6f | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-01ox-0019500000-5b93d5ee5e25639470f1 | JSpectraViewer |
|
|---|
| References |
|---|
| References: | - UniProt Consortium (2011). "Ongoing and future developments at the Universal Protein Resource." Nucleic Acids Res 39:D214-D219.21051339
- Scheer, M., Grote, A., Chang, A., Schomburg, I., Munaretto, C., Rother, M., Sohngen, C., Stelzer, M., Thiele, J., Schomburg, D. (2011). "BRENDA, the enzyme information system in 2011." Nucleic Acids Res 39:D670-D676.21062828
- Herrgard, M. J., Swainston, N., Dobson, P., Dunn, W. B., Arga, K. Y., Arvas, M., Bluthgen, N., Borger, S., Costenoble, R., Heinemann, M., Hucka, M., Le Novere, N., Li, P., Liebermeister, W., Mo, M. L., Oliveira, A. P., Petranovic, D., Pettifer, S., Simeonidis, E., Smallbone, K., Spasic, I., Weichart, D., Brent, R., Broomhead, D. S., Westerhoff, H. V., Kirdar, B., Penttila, M., Klipp, E., Palsson, B. O., Sauer, U., Oliver, S. G., Mendes, P., Nielsen, J., Kell, D. B. (2008). "A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology." Nat Biotechnol 26:1155-1160.18846089
- Lamb, D. C., Kelly, D. E., Manning, N. J., Kaderbhai, M. A., Kelly, S. L. (1999). "Biodiversity of the P450 catalytic cycle: yeast cytochrome b5/NADH cytochrome b5 reductase complex efficiently drives the entire sterol 14-demethylation (CYP51) reaction." FEBS Lett 462:283-288.10622712
|
|---|
| Synthesis Reference: | Ruan B; Wilson W K; Schroepfer G J Jr An alternative synthesis of 4,4-dimethyl-5 alpha-cholesta-8,14,24-trien-3 beta-ol, an intermediate in sterol biosynthesis and a reported activator of meiosis and of nuclear orphan receptor LXR alpha. Bioorganic & |
|---|
| External Links: | |
|---|

