| Identification |
|---|
| YMDB ID | YMDB00190 |
|---|
| Name | Sphingosine |
|---|
| Species | Saccharomyces cerevisiae |
|---|
| Strain | Baker's yeast |
|---|
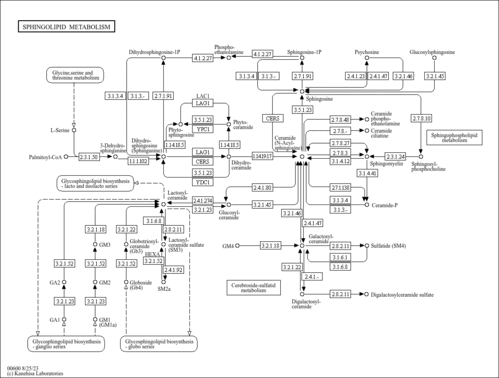
| Description | Sphingosine, also known as (4E)-sphingenine, belongs to the class of organic compounds known as 1,2-aminoalcohols. These are organic compounds containing an alkyl chain with an amine group bound to the C1 atom and an alcohol group bound to the C2 atom. Sphingosine is a very strong basic compound (based on its pKa). Sphingosine exists in all eukaryotes, ranging from yeast to humans. Within yeast, sphingosine participates in a number of enzymatic reactions. In particular, sphingosine can be converted into sphingosine 1-phosphate through its interaction with the enzyme sphingosine kinase 2. In addition, sphingosine can be biosynthesized from sphingosine 1-phosphate through the action of the enzyme sphingosine-1-phosphate phosphatase 2. In yeast, sphingosine is involved in the metabolic pathway called the sphingolipid metabolism pathway. |
|---|
| Structure | |
|---|
| Synonyms | - (-)-D-erythro-Sphingosine
- (2S,3R,4E)-2-Amino-4-octadecene-1,3-diol
- (2S,3R)-Sphingosine
- (4E)-Sphingenine
- [R-[R*,S*-(E)]]-2-amino-4-Octadecene-1,3-diol
- 4-Sphingenine
- 4-trans-Sphingenine
|
|---|
| CAS number | 123-78-4 |
|---|
| Weight | Average: 299.4919
Monoisotopic: 299.282429433 |
|---|
| InChI Key | WWUZIQQURGPMPG-CCEZHUSRSA-N |
|---|
| InChI | InChI=1S/C18H37NO2/c1-2-3-4-5-6-7-8-9-10-11-12-13-14-15-18(21)17(19)16-20/h14-15,17-18,20-21H,2-13,16,19H2,1H3/b15-14+ |
|---|
| IUPAC Name | (4E)-2-aminooctadec-4-ene-1,3-diol |
|---|
| Traditional IUPAC Name | sphingosine |
|---|
| Chemical Formula | C18H37NO2 |
|---|
| SMILES | [H]OC([H])([H])C([H])(N([H])[H])C([H])(O[H])C([H])=C([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])[H] |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as 1,2-aminoalcohols. These are organic compounds containing an alkyl chain with an amine group bound to the C1 atom and an alcohol group bound to the C2 atom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic nitrogen compounds |
|---|
| Class | Organonitrogen compounds |
|---|
| Sub Class | Amines |
|---|
| Direct Parent | 1,2-aminoalcohols |
|---|
| Alternative Parents | |
|---|
| Substituents | - Secondary alcohol
- 1,2-aminoalcohol
- Organic oxygen compound
- Organopnictogen compound
- Hydrocarbon derivative
- Primary amine
- Primary alcohol
- Organooxygen compound
- Primary aliphatic amine
- Alcohol
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Charge | 0 |
|---|
| Melting point | 81 °C |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Water Solubility | Not Available | PhysProp | | LogP | Not Available | PhysProp |
|
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations | - Nuclear Envelope, Plasma Membrane
|
|---|
| Organoleptic Properties | Not Available |
|---|
| SMPDB Pathways | |
|---|
| KEGG Pathways | |
|---|
| SMPDB Reactions | |
|---|
| KEGG Reactions | Not Available |
|---|
| Concentrations |
|---|
| Intracellular Concentrations | Not Available |
|---|
| Extracellular Concentrations | Not Available |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-03dr-9260000000-2ff04e0afd9e8b2f0851 | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-003r-8915100000-51f94df9cbd9e00dbf76 | JSpectraViewer | | LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-001i-0090000000-67307f0b7f6f858af400 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-0a59-9010000000-e66065dfabaedfa9a7f2 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-0api-9000000000-a5c7c01555839f61ab3d | JSpectraViewer | MoNA | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0f89-0092000000-c0188d9e3166e42d0a72 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-02ai-2390000000-23fd78f38dca1695f653 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4m-6940000000-bd71a7546dbbc9a6e83b | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-0090000000-310f8ec2f529dd63e5b7 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-07d4-4090000000-3ec929924c6bbc7293e0 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9030000000-c344fe4fe9af07abf710 | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 2D NMR | [1H,13C] 2D NMR Spectrum | Not Available | JSpectraViewer |
|
|---|
| References |
|---|
| References: | - UniProt Consortium (2011). "Ongoing and future developments at the Universal Protein Resource." Nucleic Acids Res 39:D214-D219.21051339
- Scheer, M., Grote, A., Chang, A., Schomburg, I., Munaretto, C., Rother, M., Sohngen, C., Stelzer, M., Thiele, J., Schomburg, D. (2011). "BRENDA, the enzyme information system in 2011." Nucleic Acids Res 39:D670-D676.21062828
|
|---|
| Synthesis Reference: | Obayashi, Michio; Schlosser, Manfred. An efficient synthesis of (2S,3R)- and (2S,3S)-sphingosine. Chemistry Letters (1985), (11), 1715-18. |
|---|
| External Links: | |
|---|