| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-000i-8920000000-c9c1de401e53d5259b37 | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (3 TMS) - 70eV, Positive | splash10-004i-7439400000-61898d5da9dfc83487d0 | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | JSpectraViewer | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF 10V, positive | splash10-0006-0900000000-b2b9deccbc523c6d0eab | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF 20V, positive | splash10-0006-0900000000-c777d18079ce5f3f7cce | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF 40V, positive | splash10-000f-9100000000-ce6e11131e810ac225a4 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - QTOF 3V, negative | splash10-002r-0910000000-f6fbf6f220b47bb44d07 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - QTOF 4V, negative | splash10-000i-0900000000-73859439e65b3b81c419 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - QTOF 5V, negative | splash10-000i-0900000000-a7d9ad79c08c938f641d | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - QTOF 7V, negative | splash10-000i-0900000000-3b8ca1c03a2b18dcb3ee | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - QTOF 10V, negative | splash10-000i-1900000000-7cb1f442b1b4017f231c | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - QTOF 15V, negative | splash10-000i-5900000000-ab5c560af44c3e113890 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - QTOF 17V, negative | splash10-000l-8900000000-188280d32d1c753f7e22 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - QTOF 20V, negative | splash10-000l-9600000000-d42db1d206b7d8fe2555 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - QTOF 23V, negative | splash10-000f-9200000000-e80c9b91e42c7ce5f07c | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - QTOF 25V, negative | splash10-0006-9200000000-88dbca436a340239632c | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - QTOF 27V, negative | splash10-0006-9100000000-9202017d9ac4e164e0df | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - QTOF 30V, negative | splash10-0006-9000000000-50c145d43aba50ec736f | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 1V, negative | splash10-0570-0940000000-573c1187a2d4556bd2c1 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 1V, negative | splash10-0570-0930000000-fb6a26ac0db1fa8e3a5f | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 2V, negative | splash10-004r-0920000000-5085342dabe01fc8bcab | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 2V, negative | splash10-004r-0910000000-1ba87cb3a1e14ed46c1a | JSpectraViewer | MoNA | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0a4i-1490000000-0c0be827ab9167718ec0 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a4r-2920000000-70cd182f84cfcb7bcc5c | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-052r-6900000000-4c4eaa55fb7ee94f5725 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-1590000000-38a825366167edaea080 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4r-2920000000-e4d23a3d35bdaecc1db4 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-06rl-6900000000-c88f4ed01c10938fc50a | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer |
|
|---|
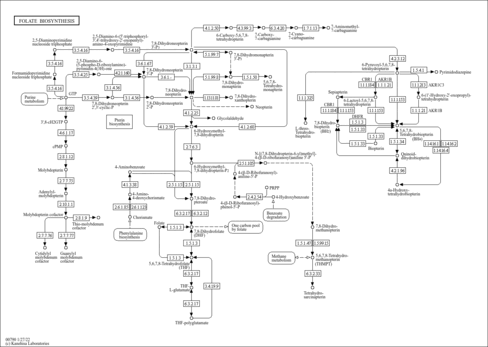
| References: | - UniProt Consortium (2011). "Ongoing and future developments at the Universal Protein Resource." Nucleic Acids Res 39:D214-D219.21051339
- Scheer, M., Grote, A., Chang, A., Schomburg, I., Munaretto, C., Rother, M., Sohngen, C., Stelzer, M., Thiele, J., Schomburg, D. (2011). "BRENDA, the enzyme information system in 2011." Nucleic Acids Res 39:D670-D676.21062828
- Herrgard, M. J., Swainston, N., Dobson, P., Dunn, W. B., Arga, K. Y., Arvas, M., Bluthgen, N., Borger, S., Costenoble, R., Heinemann, M., Hucka, M., Le Novere, N., Li, P., Liebermeister, W., Mo, M. L., Oliveira, A. P., Petranovic, D., Pettifer, S., Simeonidis, E., Smallbone, K., Spasic, I., Weichart, D., Brent, R., Broomhead, D. S., Westerhoff, H. V., Kirdar, B., Penttila, M., Klipp, E., Palsson, B. O., Sauer, U., Oliver, S. G., Mendes, P., Nielsen, J., Kell, D. B. (2008). "A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology." Nat Biotechnol 26:1155-1160.18846089
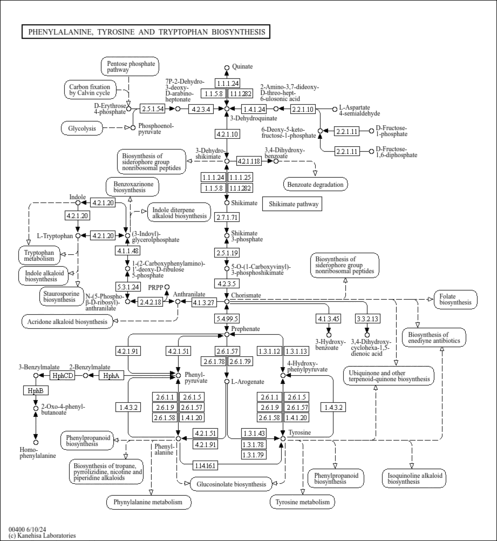
- Miozzari, G., Niederberger, P., Hutter, R. (1978). "Tryptophan biosynthesis in Saccharomyces cerevisiae: control of the flux through the pathway." J Bacteriol 134:48-59.348687
- Braus, G. H. (1991). "Aromatic amino acid biosynthesis in the yeast Saccharomyces cerevisiae: a model system for the regulation of a eukaryotic biosynthetic pathway." Microbiol Rev 55:349-370.1943992
- Henstrand, J. M., Schaller, A., Braun, M., Amrhein, N., Schmid, J. (1996). "Saccharomyces cerevisiae chorismate synthase has a flavin reductase activity." Mol Microbiol 22:859-866.8971708
- Crombie, T., Boyle, J. P., Coggins, J. R., Brown, A. J. (1994). "The folding of the bifunctional TRP3 protein in yeast is influenced by a translational pause which lies in a region of structural divergence with Escherichia coli indoleglycerol-phosphate synthase." Eur J Biochem 226:657-664.8001582
- Ball, S. G., Wickner, R. B., Cottarel, G., Schaus, M., Tirtiaux, C. (1986). "Molecular cloning and characterization of ARO7-OSM2, a single yeast gene necessary for chorismate mutase activity and growth in hypertonic medium." Mol Gen Genet 205:326-330.3027508
|
|---|