| Identification |
|---|
| YMDB ID | YMDB00075 |
|---|
| Name | Dethiobiotin |
|---|
| Species | Saccharomyces cerevisiae |
|---|
| Strain | Baker's yeast |
|---|
| Description | Dethiobiotin is an intermediate in biotin biosynthesis from 7-keto-8-aminopelargonate pathway. Biotin is a water soluble, heterocyclic cofactor for a small number of enzymes that facilitate the transfer of CO2 during carboxylation, decarboxylation, and transcarboxylation reactions involved in fatty acid and carbohydrate metabolism. Biotin biosynthesis is found in plants, bacteria, and certain fungi. [Biocyc PWY0-1507] |
|---|
| Structure | |
|---|
| Synonyms | - Desthiobiotin
- (+)-Dethiobiotin
- (4R,5S)-5-Methyl-2-oxo-4-imidazolidinehexanoic acid
- (4R-cis)-5-Methyl-2-oxo-4-imidazolidinehexanoic acid
- D-Dethiobiotin
- (4R,5S)-5-Methyl-2-oxo-4-imidazolidinehexanoate
- (4R-cis)-5-Methyl-2-oxo-4-imidazolidinehexanoate
- Desthiobiotin, (4R-cis)-isomer
- Desthiobiotin, (cis)-(+-)-isomer
- 4-Methyl-5-(omega-carboxyamyl)imidazolidone-2
- 4-Methyl-5-(ω-carboxyamyl)imidazolidone-2
- 5-Methyl-2-oxo-4-imidazolidinecaproic acid
- Dethiobiotin
- epsilon-(4-Methyl-5-imidazolidone-2)caproic acid
- ε-(4-Methyl-5-imidazolidone-2)caproic acid
|
|---|
| CAS number | 533-48-2 |
|---|
| Weight | Average: 214.2615
Monoisotopic: 214.131742452 |
|---|
| InChI Key | AUTOLBMXDDTRRT-JGVFFNPUSA-N |
|---|
| InChI | InChI=1S/C10H18N2O3/c1-7-8(12-10(15)11-7)5-3-2-4-6-9(13)14/h7-8H,2-6H2,1H3,(H,13,14)(H2,11,12,15)/t7-,8+/m0/s1 |
|---|
| IUPAC Name | 6-[(4R,5S)-5-methyl-2-oxoimidazolidin-4-yl]hexanoic acid |
|---|
| Traditional IUPAC Name | (4R,5S)-dethiobiotin |
|---|
| Chemical Formula | C10H18N2O3 |
|---|
| SMILES | [H]OC(=O)C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])[C@@]1([H])N([H])C(=O)N([H])[C@@]1([H])C([H])([H])[H] |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as medium-chain fatty acids. These are fatty acids with an aliphatic tail that contains between 4 and 12 carbon atoms. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Fatty Acyls |
|---|
| Sub Class | Fatty acids and conjugates |
|---|
| Direct Parent | Medium-chain fatty acids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Medium-chain fatty acid
- Amino fatty acid
- Heterocyclic fatty acid
- Imidazolidinone
- Imidazolidine
- Carbonic acid derivative
- Urea
- Carboxylic acid derivative
- Carboxylic acid
- Monocarboxylic acid or derivatives
- Azacycle
- Organoheterocyclic compound
- Carbonyl group
- Organic oxide
- Organopnictogen compound
- Organic oxygen compound
- Hydrocarbon derivative
- Organonitrogen compound
- Organooxygen compound
- Organic nitrogen compound
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Charge | 0 |
|---|
| Melting point | 157 °C |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Water Solubility | Not Available | PhysProp | | LogP | Not Available | PhysProp |
|
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations | |
|---|
| Organoleptic Properties | Not Available |
|---|
| SMPDB Pathways | |
|---|
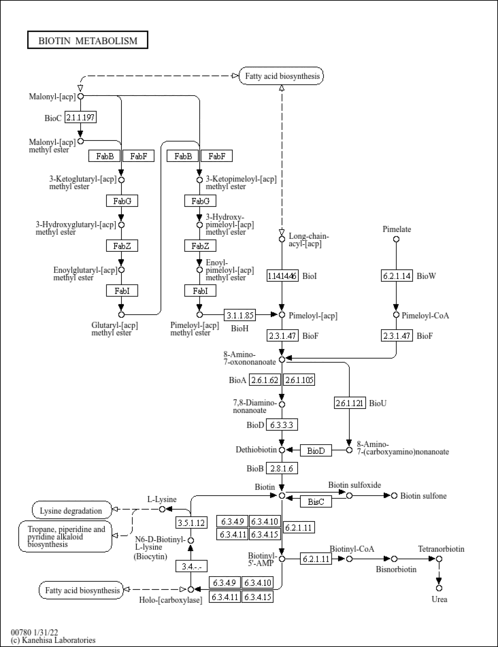
| KEGG Pathways | |
|---|
| SMPDB Reactions | |
|---|
| KEGG Reactions | |
|---|
| Concentrations |
|---|
| Intracellular Concentrations | Not Available |
|---|
| Extracellular Concentrations | Not Available |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| GC-MS | GC-MS Spectrum - GC-MS (3 TMS) | splash10-0006-3790100000-c94f58473d108e2927a3 | JSpectraViewer | MoNA | | GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-0006-1690000000-1bda89bfc3811fd0846b | JSpectraViewer | MoNA | | GC-MS | GC-MS Spectrum - GC-MS (Non-derivatized) | splash10-0006-3790100000-c94f58473d108e2927a3 | JSpectraViewer | MoNA | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0002-9500000000-0f385ef69a9d53ba7904 | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-00ba-9200000000-19b608dc5ef3430fb0b4 | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | JSpectraViewer | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-qTof , Positive | splash10-056s-4910000000-2e5662744c4e67149ef8 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-004i-9200000000-03c3baa9836a6b78a20b | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-004i-9100000000-b90f330888cd6165f177 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-056r-9000000000-7330a6fde54cd5afd3d9 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-0a4i-9000000000-42126ce46ca1e187b36a | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-IT , positive | splash10-0002-0900000000-2f9eaf52d9aa20dfeb5d | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - , positive | splash10-056s-4910000000-2e5662744c4e67149ef8 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - 35V, Positive | splash10-0002-0900000000-d8e78a6d347413a0f682 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - 10V, Positive | splash10-0002-0910000000-b2dd4a92e38e38ef8a7c | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - 40V, Positive | splash10-066r-9000000000-b2eb519e2e0e5ad91911 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-002b-4900000000-cf863dd0f0baea9dbe85 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-0006-9210000000-a459410fa0babb81e6b9 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9000000000-178bc9c95d917ad3ea55 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-03di-1190000000-d73931a407641bc332ca | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - 35V, Negative | splash10-03di-0490000000-ae866fa9d32aecb5a67d | JSpectraViewer | MoNA | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00kb-0920000000-12242f12c8a3476fe52c | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-014j-1910000000-bca8247f7dffe184f262 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00kf-9100000000-a1befeec39f013c667cc | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03di-0490000000-fbb5b60d4417d32465c9 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0006-9520000000-9247557ea0c296d0d2ad | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9000000000-a9e635f12ade363f1ca8 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03di-1090000000-7406e098948885bf0fa7 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-01ox-5930000000-26072ff1fff5e748d959 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9200000000-47fd1748a2d47287944d | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014j-0980000000-f238d4cd8c859f72d3bc | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer |
|
|---|
| References |
|---|
| References: | - UniProt Consortium (2011). "Ongoing and future developments at the Universal Protein Resource." Nucleic Acids Res 39:D214-D219.21051339
- Herrgard, M. J., Swainston, N., Dobson, P., Dunn, W. B., Arga, K. Y., Arvas, M., Bluthgen, N., Borger, S., Costenoble, R., Heinemann, M., Hucka, M., Le Novere, N., Li, P., Liebermeister, W., Mo, M. L., Oliveira, A. P., Petranovic, D., Pettifer, S., Simeonidis, E., Smallbone, K., Spasic, I., Weichart, D., Brent, R., Broomhead, D. S., Westerhoff, H. V., Kirdar, B., Penttila, M., Klipp, E., Palsson, B. O., Sauer, U., Oliver, S. G., Mendes, P., Nielsen, J., Kell, D. B. (2008). "A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology." Nat Biotechnol 26:1155-1160.18846089
- Muhlenhoff, U., Gerl, M. J., Flauger, B., Pirner, H. M., Balser, S., Richhardt, N., Lill, R., Stolz, J. (2007). "The ISC [corrected] proteins Isa1 and Isa2 are required for the function but not for the de novo synthesis of the Fe/S clusters of biotin synthase in Saccharomyces cerevisiae." Eukaryot Cell 6:495-504.17259550
- Wu, H., Ito, K., Shimoi, H. (2005). "Identification and characterization of a novel biotin biosynthesis gene in Saccharomyces cerevisiae." Appl Environ Microbiol 71:6845-6855.16269718
|
|---|
| Synthesis Reference: | Kuzuhara, Hiroyoshi; Ohrui, Hiroshi; Emoto, Sakae. Syntheses with azido sugars. II. Conversion of D-glucose to (+)-dethiobiotin. Agricultural and Biological Chemistry (1971), 35(1), 8-17. |
|---|
| External Links: | |
|---|