Hydroxymethylpyrimidine/phosphomethylpyrimidine kinase THI21 (Q08975)
| Identification | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Name | Hydroxymethylpyrimidine/phosphomethylpyrimidine kinase THI21 | ||||||||||||||||||||||||
| Synonyms |
| ||||||||||||||||||||||||
| Gene Name | THI21 | ||||||||||||||||||||||||
| Enzyme Class | |||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||
| General Function | Involved in phosphomethylpyrimidine kinase activity | ||||||||||||||||||||||||
| Specific Function | Catalyzes the phosphorylation of hydroxymethylpyrimidine phosphate (HMP-P) to HMP-PP, and also probaby that of HMP to HMP- P | ||||||||||||||||||||||||
| Cellular Location | Not Available | ||||||||||||||||||||||||
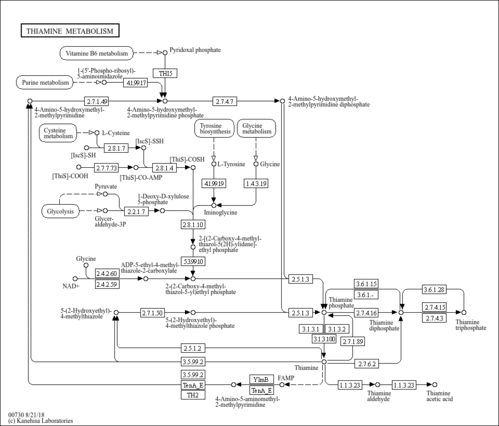
| SMPDB Pathways |
| ||||||||||||||||||||||||
| KEGG Pathways |
| ||||||||||||||||||||||||
| SMPDB Reactions | |||||||||||||||||||||||||
| KEGG Reactions | |||||||||||||||||||||||||
| Metabolites |
| ||||||||||||||||||||||||
| GO Classification |
| ||||||||||||||||||||||||
| Gene Properties | |||||||||||||||||||||||||
| Chromosome Location | chromosome 16 | ||||||||||||||||||||||||
| Locus | YPL258C | ||||||||||||||||||||||||
| Gene Sequence | >1656 bp ATGACCTATTCTACAGTTAATATTAATACACCTCCACCATATCTCGCTTTGGCCAGCAAT GAAAAATTACCCACAGTTTTGTCTATTGCTGGAACAGATCCAAGCGGTGGTGCTGGTGTT GAAGCAGATGTGAAAACTATCACTGCACACAGATGCTATGCTATGACATGCATCACTGCT TTGAATGCTCAAACTCCAGTTAAAGTATACAGCATTAATAACACGCCAAAGGAAGTAGTT TCCCAAATTTTGGACGCCAATTTACAAGACATGAAATGTGACGTCATCAAGACTGGCATG CTTACGACAGCTGCTATCGAGGTTTTGCACGAAAAACTGCTGCAGCTCGGCGAAAATAGA CCTAAGCTAGTAGTTGACCCAGTCCTTGTTGCTACTTCAGGCTCTTCTTTAGCTGGAAAG GATATAGCCAGTTTAATTACGGAGAAAATCGCACCTTTTGCTGACATTTTGACTCCTAAC ATTCCGGAGTGTTTCAAACTGTTAGGTGAAGACAGAGAAATAAGCAAATTGCGAGACATT TTCGAAGTCGCAAAAGACCTTGCCAAAATCACCAAGTGTTCCAATATTTTGGTTAAAGGT GGTCACATTCCATGGAATGATGAAGAAGGAAAGTACATCACTGATGTCCTTTACTTAGGG GCCGAACAAAGGTTTATAACCTTCAAGGGTAATTTTGTTAATACAACCCACACTCATGGT ACCGGGTGCACCCTAGCTTCCGCCATTGCATCAAATTTGGCTCGCGGTTATTCCCTTCCC CAATCCGTTTATGGCGGTATTGAGTACGTCCAAAATGCGGTGGCCATTGGCTGCGATGTC ACGAAAGAGACTGTTAAGGACAACGGGCCAATTAATCATGTCTACGCCATCGAAATTCCA TTAGAAAAAATGCTCAGTGACGAATGTTTCACTGCATCAGATGCCGTACACAAAAAGCCA GTCAAAAGTTCTCTTAACAAAATCCCTGGAGGAAGTTTCTATAAATATCTGATAAATCAC CCTAAAGTTAAGCCCCACTGGGACTCCTATGTGAACCATGATTTTGTTAGAAAAGTAGCA GACGGTTCCTTAGAGCCTAAGAAGTTCCAATTTTTTATCGAACAAGATTACCTGTACTTA GTCAATTATGCCAGGATTTCCTGCATCGCGGGTAGTAAATCCCCGTGTCTTGAAGACCTG GAGAAAGAGCTAGTTATTGTCGAATGCGTTCGTAATGGGCTTTGCCAACATGAAAGGAGA TTAAGGGAGGAATTTGGAATTAAGGATCCCGACTACTTACAAAAGATTCAAAGAGGTCCT GCATTAAGGGCTTACTGTCGTTACTTTAATGATGTTTCCAGAAGAGGTAACTGGCAGGAG TTAGTTATAGCTCTTAATCCTTGCTTAATGGGCTATGTTCACGCTTTGACCAAGATCAAG GATGAAGTAACTGCAGCAGAAGGCTCCGTTTACCGTGAATGGTGTGAAACTTACTCATCG TCATGGTGCCACGAAGCTATGCTTGAAGGCGAAAAGCTGTTGAATCACATCTTGGAAACA TACCCTCCAGAAAAACTTGATACACTGGTCACAATATATGCTGAAGTTTGTGAATTGGAA GCTAACTTCTGGACCGCCGCTCTAGAATATGAATGA | ||||||||||||||||||||||||
| Protein Properties | |||||||||||||||||||||||||
| Pfam Domain Function | |||||||||||||||||||||||||
| Protein Residues | 551 | ||||||||||||||||||||||||
| Protein Molecular Weight | 61333.69922 | ||||||||||||||||||||||||
| Protein Theoretical pI | 5.77 | ||||||||||||||||||||||||
| Signalling Regions |
| ||||||||||||||||||||||||
| Transmembrane Regions |
| ||||||||||||||||||||||||
| Protein Sequence | >Hydroxymethylpyrimidine/phosphomethylpyrimidine kinase THI21 MTYSTVNINTPPPYLALASNEKLPTVLSIAGTDPSGGAGVEADVKTITAHRCYAMTCITA LNAQTPVKVYSINNTPKEVVSQILDANLQDMKCDVIKTGMLTTAAIEVLHEKLLQLGENR PKLVVDPVLVATSGSSLAGKDIASLITEKIAPFADILTPNIPECFKLLGEDREISKLRDI FEVAKDLAKITKCSNILVKGGHIPWNDEEGKYITDVLYLGAEQRFITFKGNFVNTTHTHG TGCTLASAIASNLARGYSLPQSVYGGIEYVQNAVAIGCDVTKETVKDNGPINHVYAIEIP LEKMLSDECFTASDAVHKKPVKSSLNKIPGGSFYKYLINHPKVKPHWDSYVNHDFVRKVA DGSLEPKKFQFFIEQDYLYLVNYARISCIAGSKSPCLEDLEKELVIVECVRNGLCQHERR LREEFGIKDPDYLQKIQRGPALRAYCRYFNDVSRRGNWQELVIALNPCLMGYVHALTKIK DEVTAAEGSVYREWCETYSSSWCHEAMLEGEKLLNHILETYPPEKLDTLVTIYAEVCELE ANFWTAALEYE | ||||||||||||||||||||||||
| References | |||||||||||||||||||||||||
| External Links |
| ||||||||||||||||||||||||
| General Reference |
| ||||||||||||||||||||||||