Putative dehydrodolichyl diphosphate synthase (Q03175)
| Identification | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Name | Putative dehydrodolichyl diphosphate synthase | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene Name | SRT1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Enzyme Class | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| General Function | Involved in transferase activity, transferring alkyl or aryl (other than methyl) groups | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Specific Function | Could be a cis-prenyl transferase that adds multiple copies of isopentenyl pyrophosphate (ipp) to farnesyl pyrophosphate (FPP) to produce dehydrodolichyl diphosphate (DeDol- PP) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Location | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMPDB Pathways |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
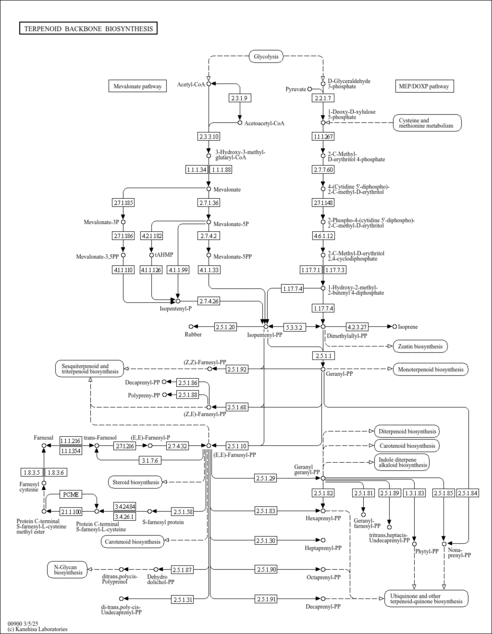
| KEGG Pathways |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMPDB Reactions |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG Reactions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolites |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| GO Classification |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chromosome Location | chromosome 13 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Locus | YMR101C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene Sequence | >1032 bp ATGAAAATGCCCAGTATTATTCAGATTCAGTTTGTAGCCCTAAAAAGGCTTTTGGTAGAA ACCAAAGAACAGATGTGCTTCGCAGTGAAAAGTATATTTCAGAGAGTATTTGCGTGGGTT ATGTCATTAAGCTTGTTTTCATGGTTTTATGTAAATCTTCAGAATATTTTGATAAAAGCA TTAAGGGTAGGGCCAGTGCCTGAACATGTCTCCTTTATCATGGATGGTAACCGGAGATAT GCCAAGTCAAGAAGGCTACCAGTAAAAAAAGGCCATGAAGCTGGTGGGTTAACGTTACTA ACACTACTGTATATCTGCAAAAGATTGGGTGTAAAATGTGTTTCCGCCTATGCATTTTCT ATTGAAAATTTTAATAGACCAAAAGAAGAAGTAGATACGCTAATGAATTTGTTTACGGTA AAGCTTGATGAATTCGCAAAAAGAGCCAAGGACTATAAGGATCCCTTATACGGATCTAAA ATAAGAATAGTAGGTGATCAATCTTTACTATCTCCAGAAATGAGAAAAAAAATTAAAAAA GTGGAAGAAATCACACAGGATGGAGACGATTTCACTTTATTTATATGTTTTCCTTACACT TCAAGAAATGATATGTTACATACTATTCGTGATTCAGTTGAAGACCATTTGGAAAATAAA TCACCAAGGATTAATATAAGAAAATTTACTAATAAAATGTACATGGGTTTCCATTCCAAT AAATGTGAATTATTAATCAGAACAAGTGGGCATAGGAGGCTCTCAGACTATATGCTATGG CAAGTACATGAAAATGCCACCATTGAATTTAGTGATACGTTGTGGCCAAATTTTAGCTTC TTTGCTATGTACCTGATGATTCTCAAATGGTCCTTCTTTTCCACCATTCAAAAATATAAT GAGAAGAATCACTCATTGTTTGAAAAAATACATGAAAGCGTTCCTTCAATATTTAAAAAA AAGAAAACAGCTATGTCTTTGTACAACTTTCCAAACCCCCCCATTTCAGTTTCGGTTACA GGAGATGAATAA | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Protein Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pfam Domain Function |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Protein Residues | 343 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Protein Molecular Weight | 40199.69922 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Protein Theoretical pI | 10.03 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Signalling Regions |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Transmembrane Regions |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Protein Sequence | >Putative dehydrodolichyl diphosphate synthase MKMPSIIQIQFVALKRLLVETKEQMCFAVKSIFQRVFAWVMSLSLFSWFYVNLQNILIKA LRVGPVPEHVSFIMDGNRRYAKSRRLPVKKGHEAGGLTLLTLLYICKRLGVKCVSAYAFS IENFNRPKEEVDTLMNLFTVKLDEFAKRAKDYKDPLYGSKIRIVGDQSLLSPEMRKKIKK VEEITQDGDDFTLFICFPYTSRNDMLHTIRDSVEDHLENKSPRINIRKFTNKMYMGFHSN KCELLIRTSGHRRLSDYMLWQVHENATIEFSDTLWPNFSFFAMYLMILKWSFFSTIQKYN EKNHSLFEKIHESVPSIFKKKKTAMSLYNFPNPPISVSVTGDE | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| General Reference |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||