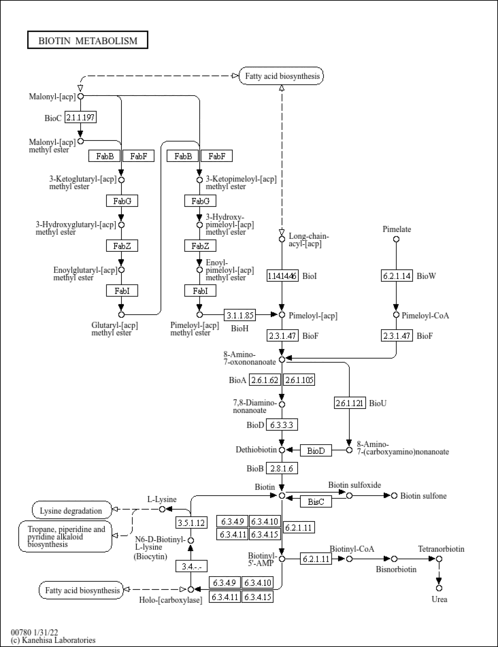
| General Reference | - Phalip, V., Kuhn, I., Lemoine, Y., Jeltsch, J. M. (1999). "Characterization of the biotin biosynthesis pathway in Saccharomyces cerevisiae and evidence for a cluster containing BIO5, a novel gene involved in vitamer uptake." Gene 232:43-51.10333520
- Wu, H., Ito, K., Shimoi, H. (2005). "Identification and characterization of a novel biotin biosynthesis gene in Saccharomyces cerevisiae." Appl Environ Microbiol 71:6845-6855.16269718
- Philippsen, P., Kleine, K., Pohlmann, R., Dusterhoft, A., Hamberg, K., Hegemann, J. H., Obermaier, B., Urrestarazu, L. A., Aert, R., Albermann, K., Altmann, R., Andre, B., Baladron, V., Ballesta, J. P., Becam, A. M., Beinhauer, J., Boskovic, J., Buitrago, M. J., Bussereau, F., Coster, F., Crouzet, M., D'Angelo, M., Dal Pero, F., De Antoni, A., Del Rey, F., Doignon, F., Domdey, H., Dubois, E., Fiedler, T., Fleig, U., Floeth, M., Fritz, C., Gaillardin, C., Garcia-Cantalejo, J. M., Glansdorff, N. N., Goffeau, A., Gueldener, U., Herbert, C., Heumann, K., Heuss-Neitzel, D., Hilbert, H., Hinni, K., Iraqui Houssaini, I., Jacquet, M., Jimenez, A., Jonniaux, J. L., Karpfinger, L., Lanfranchi, G., Lepingle, A., Levesque, H., Lyck, R., Maftahi, M., Mallet, L., Maurer, K. C., Messenguy, F., Mewes, H. W., Mosti, D., Nasr, F., Nicaud, J. M., Niedenthal, R. K., Pandolfo, D., Pierard, A., Piravandi, E., Planta, R. J., Pohl, T. M., Purnelle, B., Rebischung, C., Remacha, M., Revuelta, J. L., Rinke, M., Saiz, J. E., Sartorello, F., Scherens, B., Sen-Gupta, M., Soler-Mira, A., Urbanus, J. H., Valle, G., Van Dyck, L., Verhasselt, P., Vierendeels, F., Vissers, S., Voet, M., Volckaert, G., Wach, A., Wambutt, R., Wedler, H., Zollner, A., Hani, J. (1997). "The nucleotide sequence of Saccharomyces cerevisiae chromosome XIV and its evolutionary implications." Nature 387:93-98.9169873
|
|---|