Dihydroxy-acid dehydratase, mitochondrial (P39522)
| Identification | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Name | Dihydroxy-acid dehydratase, mitochondrial | ||||||||||||||||||||||||
| Synonyms |
| ||||||||||||||||||||||||
| Gene Name | ILV3 | ||||||||||||||||||||||||
| Enzyme Class | |||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||
| General Function | Involved in metabolic process | ||||||||||||||||||||||||
| Specific Function | 2,3-dihydroxy-3-methylbutanoate = 3-methyl-2- oxobutanoate + H(2)O | ||||||||||||||||||||||||
| Cellular Location | Mitochondrion | ||||||||||||||||||||||||
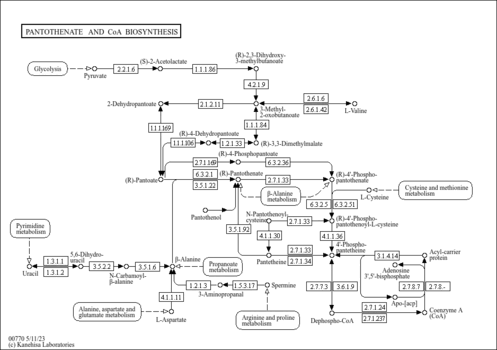
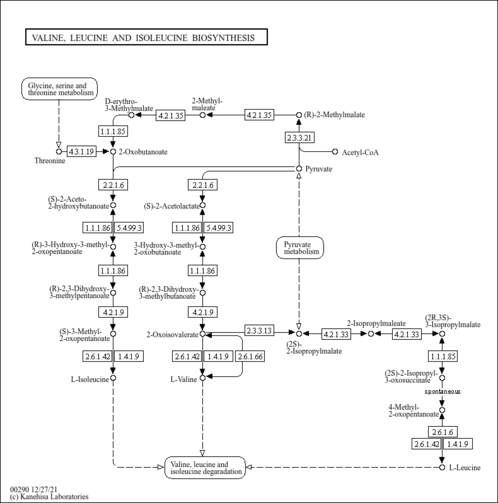
| SMPDB Pathways |
| ||||||||||||||||||||||||
| KEGG Pathways |
| ||||||||||||||||||||||||
| SMPDB Reactions | |||||||||||||||||||||||||
| KEGG Reactions |
| ||||||||||||||||||||||||
| Metabolites |
| ||||||||||||||||||||||||
| GO Classification |
| ||||||||||||||||||||||||
| Gene Properties | |||||||||||||||||||||||||
| Chromosome Location | chromosome 10 | ||||||||||||||||||||||||
| Locus | YJR016C | ||||||||||||||||||||||||
| Gene Sequence | >1758 bp ATGGGCTTGTTAACGAAAGTTGCTACATCTAGACAATTCTCTACAACGAGATGCGTTGCA AAGAAGCTCAACAAGTACTCGTATATCATCACTGAACCTAAGGGCCAAGGTGCGTCCCAG GCCATGCTTTATGCCACCGGTTTCAAGAAGGAAGATTTCAAGAAGCCTCAAGTCGGGGTT GGTTCCTGTTGGTGGTCCGGTAACCCATGTAACATGCATCTATTGGACTTGAATAACAGA TGTTCTCAATCCATTGAAAAAGCGGGTTTGAAAGCTATGCAGTTCAACACCATCGGTGTT TCAGACGGTATCTCTATGGGTACTAAAGGTATGAGATACTCGTTACAAAGTAGAGAAATC ATTGCAGACTCCTTTGAAACCATCATGATGGCACAACACTACGATGCTAACATCGCCATC CCATCATGTGACAAAAACATGCCCGGTGTCATGATGGCCATGGGTAGACATAACAGACCT TCCATCATGGTATATGGTGGTACTATCTTGCCCGGTCATCCAACATGTGGTTCTTCGAAG ATCTCTAAAAACATCGATATCGTCTCTGCGTTCCAATCCTACGGTGAATATATTTCCAAG CAATTCACTGAAGAAGAAAGAGAAGATGTTGTGGAACATGCATGCCCAGGTCCTGGTTCT TGTGGTGGTATGTATACTGCCAACACAATGGCTTCTGCCGCTGAAGTGCTAGGTTTGACC ATTCCAAACTCCTCTTCCTTCCCAGCCGTTTCCAAGGAGAAGTTAGCTGAGTGTGACAAC ATTGGTGAATACATCAAGAAGACAATGGAATTGGGTATTTTACCTCGTGATATCCTCACA AAAGAGGCTTTTGAAAACGCCATTACTTATGTCGTTGCAACCGGTGGGTCCACTAATGCT GTTTTGCATTTGGTGGCTGTTGCTCACTCTGCGGGTGTCAAGTTGTCACCAGATGATTTC CAAAGAATCAGTGATACTACACCATTGATCGGTGACTTCAAACCTTCTGGTAAATACGTC ATGGCCGATTTGATTAACGTTGGTGGTACCCAATCTGTGATTAAGTATCTATATGAAAAC AACATGTTGCACGGTAACACAATGACTGTTACCGGTGACACTTTGGCAGAACGTGCAAAG AAAGCACCAAGCCTACCTGAAGGACAAGAGATTATTAAGCCACTCTCCCACCCAATCAAG GCCAACGGTCACTTGCAAATTCTGTACGGTTCATTGGCACCAGGTGGAGCTGTGGGTAAA ATTACCGGTAAGGAAGGTACTTACTTCAAGGGTAGAGCACGTGTGTTCGAAGAGGAAGGT GCCTTTATTGAAGCCTTGGAAAGAGGTGAAATCAAGAAGGGTGAAAAAACCGTTGTTGTT ATCAGATATGAAGGTCCAAGAGGTGCACCAGGTATGCCTGAAATGCTAAAGCCTTCCTCT GCTCTGATGGGTTACGGTTTGGGTAAAGATGTTGCATTGTTGACTGATGGTAGATTCTCT GGTGGTTCTCACGGGTTCTTAATCGGCCACATTGTTCCCGAAGCCGCTGAAGGTGGTCCT ATCGGGTTGGTCAGAGACGGCGATGAGATTATCATTGATGCTGATAATAACAAGATTGAC CTATTAGTCTCTGATAAGGAAATGGCTCAACGTAAACAAAGTTGGGTTGCACCTCCACCT CGTTACACAAGAGGTACTCTATCCAAGTATGCTAAGTTGGTTTCCAACGCTTCCAACGGT TGTGTTTTAGATGCTTGA | ||||||||||||||||||||||||
| Protein Properties | |||||||||||||||||||||||||
| Pfam Domain Function |
| ||||||||||||||||||||||||
| Protein Residues | 585 | ||||||||||||||||||||||||
| Protein Molecular Weight | 62860.60156 | ||||||||||||||||||||||||
| Protein Theoretical pI | 7.92 | ||||||||||||||||||||||||
| Signalling Regions |
| ||||||||||||||||||||||||
| Transmembrane Regions |
| ||||||||||||||||||||||||
| Protein Sequence | >Dihydroxy-acid dehydratase, mitochondrial MGLLTKVATSRQFSTTRCVAKKLNKYSYIITEPKGQGASQAMLYATGFKKEDFKKPQVGV GSCWWSGNPCNMHLLDLNNRCSQSIEKAGLKAMQFNTIGVSDGISMGTKGMRYSLQSREI IADSFETIMMAQHYDANIAIPSCDKNMPGVMMAMGRHNRPSIMVYGGTILPGHPTCGSSK ISKNIDIVSAFQSYGEYISKQFTEEEREDVVEHACPGPGSCGGMYTANTMASAAEVLGLT IPNSSSFPAVSKEKLAECDNIGEYIKKTMELGILPRDILTKEAFENAITYVVATGGSTNA VLHLVAVAHSAGVKLSPDDFQRISDTTPLIGDFKPSGKYVMADLINVGGTQSVIKYLYEN NMLHGNTMTVTGDTLAERAKKAPSLPEGQEIIKPLSHPIKANGHLQILYGSLAPGGAVGK ITGKEGTYFKGRARVFEEEGAFIEALERGEIKKGEKTVVVIRYEGPRGAPGMPEMLKPSS ALMGYGLGKDVALLTDGRFSGGSHGFLIGHIVPEAAEGGPIGLVRDGDEIIIDADNNKID LLVSDKEMAQRKQSWVAPPPRYTRGTLSKYAKLVSNASNGCVLDA | ||||||||||||||||||||||||
| References | |||||||||||||||||||||||||
| External Links |
| ||||||||||||||||||||||||
| General Reference |
| ||||||||||||||||||||||||