| Identification |
|---|
| Name | Phosphatidic acid phosphohydrolase 1 |
|---|
| Synonyms | |
|---|
| Gene Name | PAH1 |
|---|
| Enzyme Class | Not Available |
|---|
| Biological Properties |
|---|
| General Function | vacuole fusion, non-autophagic |
|---|
| Specific Function | Mg(2+)-dependent phosphatidate (PA) phosphatase which catalyzes the dephosphorylation of PA to yield diacylglycerol. Required for de novo lipid synthesis and formation of lipid droplets. Controles transcription of phospholipid biosynthetic genes and nuclear structure by regulating the amount of membrane present at the nuclear envelope. Involved in plasmid maintenance, in respiration and in cell proliferation. |
|---|
| Cellular Location | Not Available |
|---|
| SMPDB Pathways | | Triacylglycerol metabolism TG(12:0/16:0/16:1(9Z)) | PW007664 | 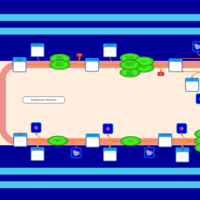  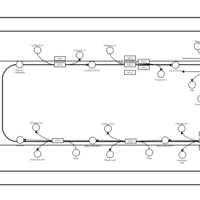 | | Triacylglycerol metabolism TG(12:0/16:0/18:0) | PW007707 | 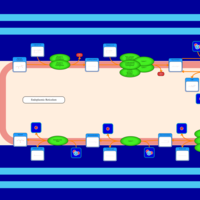 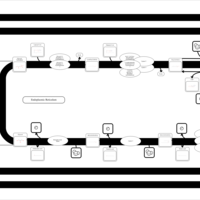  | | Triacylglycerol metabolism TG(12:0/16:0/18:1(9Z)) | PW007721 | 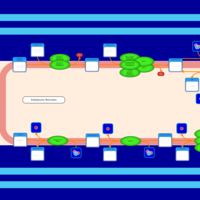   | | Triacylglycerol metabolism TG(12:0/16:0/20:0) | PW007753 | 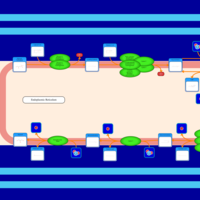 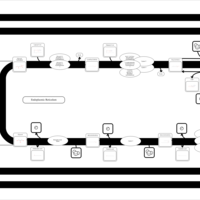 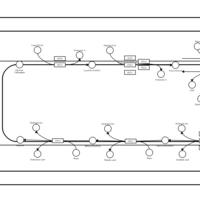 | | Triacylglycerol metabolism TG(12:0/16:0/22:0) | PW007776 | 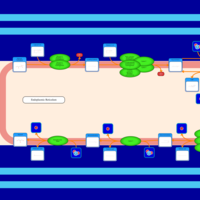 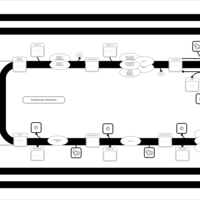 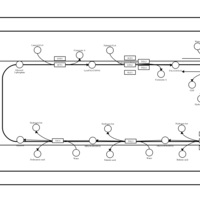 |
|
|---|
| KEGG Pathways | | Glycerophospholipid metabolism | ec00564 |  |
|
|---|
| SMPDB Reactions |
|
|---|
| KEGG Reactions | Not Available |
|---|
| Metabolites | | YMDB ID | Name | View |
|---|
| YMDB00890 | water | Show | | YMDB00907 | phosphate | Show | | YMDB01127 | DG(16:0/16:0/0:0) | Show | | YMDB01161 | PA(16:0/16:0) | Show | | YMDB12737 | DG(14:0/14:0/0:0) | Show |
|
|---|
| GO Classification | | Function |
|---|
| cytosol | | vacuole fusion, non-autophagic | | positive regulation of transcription from RNA polymerase II promoter | | transcription, DNA-templated | | nuclear membrane | | lipid particle | | extrinsic component of membrane | | vacuole | | phosphatidate phosphatase activity | | transcription coactivator activity | | transcription regulatory region DNA binding | | aerobic respiration | | fatty acid catabolic process | | lipid particle organization | | phospholipid biosynthetic process | | plasmid maintenance | | triglyceride biosynthetic process | | endoplasmic reticulum membrane |
|
|---|
| Gene Properties |
|---|
| Chromosome Location | Not Available |
|---|
| Locus | |
|---|
| Gene Sequence | >2589 bp
ATGCAGTACGTAGGCAGAGCTCTTGGGTCTGTGTCTAAAACATGGTCTTCTATCAATCCG
GCTACGCTATCAGGTGCTATAGATGTCATTGTAGTGGAGCATCCAGACGGAAGGCTATCA
TGTTCTCCCTTTCATGTGAGGTTCGGCAAATTTCAAATTCTAAAGCCATCTCAAAAGAAA
GTCCAAGTGTTTATAAATGAGAAACTGAGTAATATGCCAATGAAACTGAGTGATTCTGGA
GAAGCCTATTTCGTTTTCGAGATGGGTGACCAGGTCACTGATGTCCCTGACGAATTGCTT
GTGTCGCCCGTGATGAGCGCCACATCAAGCCCCCCTCAATCACCTGAAACATCCATCTTA
GAAGGAGGAACCGAGGGTGAAGGTGAAGGTGAAAATGAAAATAAGAAGAAGGAAAAGAAA
GTGCTAGAGGAACCAGATTTTTTAGATATCAATGACACTGGAGATTCAGGCAGTAAAAAT
AGTGAAACTACAGGGTCGCTTTCTCCTACTGAATCCTCTACAACGACACCACCAGATTCA
GTTGAAGAGAGGAAGCTTGTTGAGCAGCGTACAAAGAACTTTCAGCAAAAACTAAACAAA
AAACTCACTGAAATCCATATACCCAGTAAACTTGATAACAATGGCGACTTACTACTAGAC
ACTGAAGGTTACAAGCCAAACAAGAATATGATGCATGACACAGACATACAACTGAAGCAG
TTGTTAAAGGACGAATTCGGTAATGATTCAGATATTTCCAGTTTTATCAAGGAGGACAAA
AATGGCAACATCAAGATCGTAAATCCTTACGAGCACCTTACTGATTTATCTCCTCCAGGT
ACGCCTCCAACAATGGCCACAAGCGGATCAGTTTTAGGCTTAGATGCAATGGAATCAGGA
AGTACTTTGAATTCGTTATCTTCTTCACCTTCTGGTTCCGATACTGAGGACGAAACATCA
TTTAGCAAAGAACAAAGCAGTAAAAGTGAAAAAACTAGCAAGAAAGGAACAGCAGGGAGC
GGTGAGACCGAGAAAAGATACATACGAACGATAAGATTGACTAATGACCAGTTAAAGTGC
CTAAATTTAACTTATGGTGAAAATGATCTGAAATTTTCCGTAGATCACGGAAAAGCTATT
GTTACGTCAAAATTATTCGTTTGGAGGTGGGATGTTCCAATTGTTATCAGTGATATTGAT
GGCACCATCACAAAATCGGACGCTTTAGGCCATGTTCTGGCAATGATAGGAAAAGACTGG
ACGCACTTGGGTGTAGCCAAGTTATTTAGCGAGATCTCCAGGAATGGCTATAATATACTC
TATCTAACTGCAAGAAGTGCTGGACAAGCTGATTCCACGAGGAGTTATTTGCGATCAATT
GAACAGAATGGCAGCAAACTACCAAATGGGCCTGTGATTTTATCACCCGATAGAACGATG
GCTGCGTTAAGGCGGGAAGTAATACTAAAAAAACCTGAAGTCTTTAAAATCGCGTGTCTA
AACGACATAAGATCCTTGTATTTTGAAGACAGTGATAACGAAGTGGATACAGAGGAAAAA
TCAACACCATTTTTTGCCGGCTTTGGTAATAGGATTACTGATGCTTTATCTTACAGAACT
GTGGGGATACCTAGTTCAAGAATTTTCACAATAAATACAGAGGGTGAGGTTCATATGGAA
TTATTGGAGTTAGCAGGTTACAGAAGCTCCTATATTCATATCAATGAGCTTGTCGATCAT
TTCTTTCCACCAGTCAGCCTTGATAGTGTCGATCTAAGAACTAATACTTCCATGGTTCCT
GGCTCCCCCCCTAATAGAACGTTGGATAACTTTGACTCAGAAATTACTTCAGGTCGCAAA
ACGCTATTTAGAGGCAATCAGGAAGAGAAATTCACAGACGTAAATTTTTGGAGAGACCCG
TTAGTCGACATCGACAACTTATCGGATATTAGCAATGATGATTCTGATAACATCGATGAA
GATACTGACGTATCACAACAAAGCAACATTAGTAGAAATAGGGCAAATTCAGTCAAAACC
GCCAAGGTCACTAAAGCCCCGCAAAGAAATGTGAGCGGCAGCACAAATAACAACGAAGTT
TTAGCCGCTTCGTCTGATGTAGAAAATGCGTCTGACCTGGTGAGTTCCCATAGTAGCTCA
GGATCCACGCCCAATAAATCTACAATGTCCAAAGGGGACATTGGAAAACAAATATATTTG
GAGCTAGGTTCTCCACTTGCATCGCCAAAACTAAGATATTTAGACGATATGGATGATGAA
GACTCCAATTACAATAGAACTAAATCAAGGAGAGCATCTTCTGCAGCCGCGACTAGTATC
GATAAAGAGTTCAAAAAGCTCTCTGTGTCAAAGGCCGGCGCTCCAACAAGAATTGTTTCA
AAGATCAACGTTTCAAATGACGTACATTCACTTGGGAATTCAGATACCGAATCACGAAGG
GAGCAAAGTGTTAATGAAACAGGGCGCAATCAGCTACCCCACAACTCAATGGACGATAAA
GATTTGGATTCAAGAGTAAGCGATGAATTCGATGACGATGAATTCGACGAAGATGAATTC
GAAGATTAA |
|---|
| Protein Properties |
|---|
| Pfam Domain Function | Not Available |
|---|
| Protein Residues | 862 |
|---|
| Protein Molecular Weight | 95029.985 |
|---|
| Protein Theoretical pI | Not Available |
|---|
| Signalling Regions | Not Available |
|---|
| Transmembrane Regions | Not Available |
|---|
| Protein Sequence | >Phosphatidic acid phosphohydrolase 1
MQYVGRALGSVSKTWSSINPATLSGAIDVIVVEHPDGRLSCSPFHVRFGKFQILKPSQKK
VQVFINEKLSNMPMKLSDSGEAYFVFEMGDQVTDVPDELLVSPVMSATSSPPQSPETSIL
EGGTEGEGEGENENKKKEKKVLEEPDFLDINDTGDSGSKNSETTGSLSPTESSTTTPPDS
VEERKLVEQRTKNFQQKLNKKLTEIHIPSKLDNNGDLLLDTEGYKPNKNMMHDTDIQLKQ
LLKDEFGNDSDISSFIKEDKNGNIKIVNPYEHLTDLSPPGTPPTMATSGSVLGLDAMESG
STLNSLSSSPSGSDTEDETSFSKEQSSKSEKTSKKGTAGSGETEKRYIRTIRLTNDQLKC
LNLTYGENDLKFSVDHGKAIVTSKLFVWRWDVPIVISDIDGTITKSDALGHVLAMIGKDW
THLGVAKLFSEISRNGYNILYLTARSAGQADSTRSYLRSIEQNGSKLPNGPVILSPDRTM
AALRREVILKKPEVFKIACLNDIRSLYFEDSDNEVDTEEKSTPFFAGFGNRITDALSYRT
VGIPSSRIFTINTEGEVHMELLELAGYRSSYIHINELVDHFFPPVSLDSVDLRTNTSMVP
GSPPNRTLDNFDSEITSGRKTLFRGNQEEKFTDVNFWRDPLVDIDNLSDISNDDSDNIDE
DTDVSQQSNISRNRANSVKTAKVTKAPQRNVSGSTNNNEVLAASSDVENASDLVSSHSSS
GSTPNKSTMSKGDIGKQIYLELGSPLASPKLRYLDDMDDEDSNYNRTKSRRASSAAATSI
DKEFKKLSVSKAGAPTRIVSKINVSNDVHSLGNSDTESRREQSVNETGRNQLPHNSMDDK
DLDSRVSDEFDDDEFDEDEFED |
|---|
| References |
|---|
| External Links | | Resource | Link |
|---|
| Saccharomyces Genome Database | PAH1 | | Uniprot ID | P32567 | | Uniprot Name | PAH1 |
|
|---|
| General Reference | Not Available |
|---|
