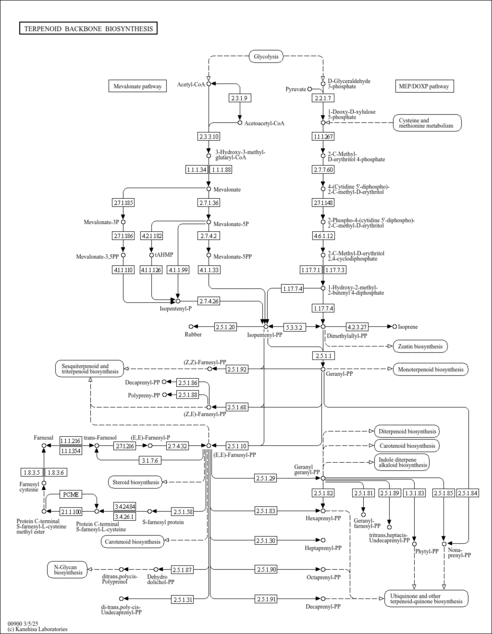
| General Reference | - Toth, M. J., Huwyler, L. (1996). "Molecular cloning and expression of the cDNAs encoding human and yeast mevalonate pyrophosphate decarboxylase." J Biol Chem 271:7895-7898.8626466
- Berges, T., Guyonnet, D., Karst, F. (1997). "The Saccharomyces cerevisiae mevalonate diphosphate decarboxylase is essential for viability, and a single Leu-to-Pro mutation in a conserved sequence leads to thermosensitivity." J Bacteriol 179:4664-4670.9244250
- Philippsen, P., Kleine, K., Pohlmann, R., Dusterhoft, A., Hamberg, K., Hegemann, J. H., Obermaier, B., Urrestarazu, L. A., Aert, R., Albermann, K., Altmann, R., Andre, B., Baladron, V., Ballesta, J. P., Becam, A. M., Beinhauer, J., Boskovic, J., Buitrago, M. J., Bussereau, F., Coster, F., Crouzet, M., D'Angelo, M., Dal Pero, F., De Antoni, A., Del Rey, F., Doignon, F., Domdey, H., Dubois, E., Fiedler, T., Fleig, U., Floeth, M., Fritz, C., Gaillardin, C., Garcia-Cantalejo, J. M., Glansdorff, N. N., Goffeau, A., Gueldener, U., Herbert, C., Heumann, K., Heuss-Neitzel, D., Hilbert, H., Hinni, K., Iraqui Houssaini, I., Jacquet, M., Jimenez, A., Jonniaux, J. L., Karpfinger, L., Lanfranchi, G., Lepingle, A., Levesque, H., Lyck, R., Maftahi, M., Mallet, L., Maurer, K. C., Messenguy, F., Mewes, H. W., Mosti, D., Nasr, F., Nicaud, J. M., Niedenthal, R. K., Pandolfo, D., Pierard, A., Piravandi, E., Planta, R. J., Pohl, T. M., Purnelle, B., Rebischung, C., Remacha, M., Revuelta, J. L., Rinke, M., Saiz, J. E., Sartorello, F., Scherens, B., Sen-Gupta, M., Soler-Mira, A., Urbanus, J. H., Valle, G., Van Dyck, L., Verhasselt, P., Vierendeels, F., Vissers, S., Voet, M., Volckaert, G., Wach, A., Wambutt, R., Wedler, H., Zollner, A., Hani, J. (1997). "The nucleotide sequence of Saccharomyces cerevisiae chromosome XIV and its evolutionary implications." Nature 387:93-98.9169873
- Hu, Y., Rolfs, A., Bhullar, B., Murthy, T. V., Zhu, C., Berger, M. F., Camargo, A. A., Kelley, F., McCarron, S., Jepson, D., Richardson, A., Raphael, J., Moreira, D., Taycher, E., Zuo, D., Mohr, S., Kane, M. F., Williamson, J., Simpson, A., Bulyk, M. L., Harlow, E., Marsischky, G., Kolodner, R. D., LaBaer, J. (2007). "Approaching a complete repository of sequence-verified protein-encoding clones for Saccharomyces cerevisiae." Genome Res 17:536-543.17322287
- Ashby, M. N., Kutsunai, S. Y., Ackerman, S., Tzagoloff, A., Edwards, P. A. (1992). "COQ2 is a candidate for the structural gene encoding para-hydroxybenzoate:polyprenyltransferase." J Biol Chem 267:4128-4136.1740455
- Ghaemmaghami, S., Huh, W. K., Bower, K., Howson, R. W., Belle, A., Dephoure, N., O'Shea, E. K., Weissman, J. S. (2003). "Global analysis of protein expression in yeast." Nature 425:737-741.14562106
- Albuquerque, C. P., Smolka, M. B., Payne, S. H., Bafna, V., Eng, J., Zhou, H. (2008). "A multidimensional chromatography technology for in-depth phosphoproteome analysis." Mol Cell Proteomics 7:1389-1396.18407956
- Bonanno, J. B., Edo, C., Eswar, N., Pieper, U., Romanowski, M. J., Ilyin, V., Gerchman, S. E., Kycia, H., Studier, F. W., Sali, A., Burley, S. K. (2001). "Structural genomics of enzymes involved in sterol/isoprenoid biosynthesis." Proc Natl Acad Sci U S A 98:12896-12901.11698677
|
|---|