3-hydroxy-3-methylglutaryl-coenzyme A reductase 2 (P12684)
| Identification | |||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Name | 3-hydroxy-3-methylglutaryl-coenzyme A reductase 2 | ||||||||||||||||||||||||||||
| Synonyms |
| ||||||||||||||||||||||||||||
| Gene Name | HMG2 | ||||||||||||||||||||||||||||
| Enzyme Class | |||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||
| General Function | Involved in hydroxymethylglutaryl-CoA reductase (NADPH) activity | ||||||||||||||||||||||||||||
| Specific Function | This transmembrane glycoprotein is involved in the control of cholesterol biosynthesis. It is the rate-limiting enzyme of the sterol biosynthesis | ||||||||||||||||||||||||||||
| Cellular Location | Endoplasmic reticulum membrane; Multi-pass membrane protein | ||||||||||||||||||||||||||||
| SMPDB Pathways |
| ||||||||||||||||||||||||||||
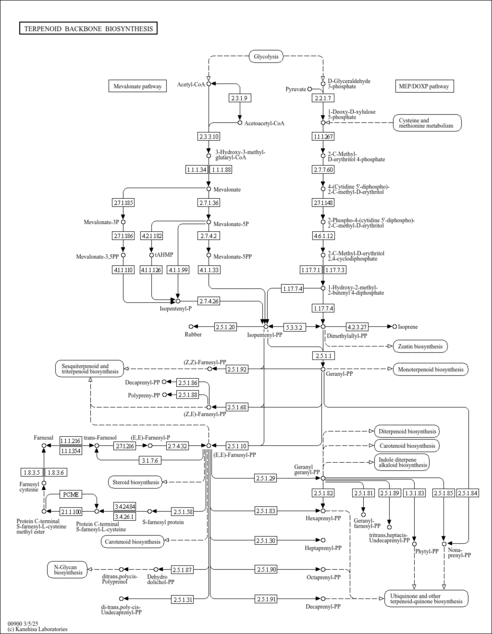
| KEGG Pathways |
| ||||||||||||||||||||||||||||
| SMPDB Reactions |
| ||||||||||||||||||||||||||||
| KEGG Reactions |
| ||||||||||||||||||||||||||||
| Metabolites |
| ||||||||||||||||||||||||||||
| GO Classification |
| ||||||||||||||||||||||||||||
| Gene Properties | |||||||||||||||||||||||||||||
| Chromosome Location | chromosome 12 | ||||||||||||||||||||||||||||
| Locus | YLR450W | ||||||||||||||||||||||||||||
| Gene Sequence | >3138 bp ATGTCACTTCCCTTAAAAACGATAGTACATTTGGTAAAGCCCTTTGCTTGCACTGCTAGG TTTAGTGCGAGATACCCAATCCACGTCATTGTTGTTGCTGTTTTATTGAGTGCCGCTGCT TATCTATCCGTGACACAATCTTACCTTAACGAATGGAAGCTGGACTCTAATCAGTATTCT ACATACTTAAGCATAAAGCCGGATGAGTTGTTTGAAAAATGCACACACTACTATAGGTCT CCTGTGTCTGATACATGGAAGTTACTCAGCTCTAAAGAAGCCGCCGATATTTATACCCCT TTTCATTATTATTTGTCTACCATAAGTTTTCAAAGTAAGGACAATTCAACGACTTTGCCT TCCCTTGATGACGTTATTTACAGTGTTGACCATACCAGGTACTTATTAAGTGAAGAGCCA AAGATACCAACTGAACTAGTGTCTGAAAACGGAACGAAATGGAGATTGAGAAACAACAGC AATTTTATTTTGGACCTGCATAATATTTACCGAAATATGGTGAAGCAATTTTCTAACAAA ACGAGCGAATTTGATCAGTTCGATTTGTTTATCATCCTAGCTGCTTACCTTACTCTTTTT TATACTCTCTGTTGCCTGTTTAATGACATGAGGAAAATCGGATCAAAGTTTTGGTTAAGC TTTTCTGCTCTTTCAAACTCTGCATGCGCATTATATTTATCGCTGTACACAACTCACAGT TTATTGAAGAAACCGGCTTCCTTATTAAGTTTGGTCATTGGACTACCATTTATCGTAGTA ATTATTGGCTTTAAGCATAAAGTTCGACTTGCGGCATTCTCGCTACAAAAATTCCACAGA ATTAGTATTGACAAGAAAATAACGGTAAGCAACATTATTTATGAGGCTATGTTTCAAGAA GGTGCCTACTTAATCCGCGACTACTTATTTTATATTAGCTCCTTCATTGGATGTGCTATT TATGCTAGACATCTTCCCGGATTGGTCAATTTCTGTATTTTGTCTACATTTATGCTAGTT TTCGACTTGCTTTTGTCTGCTACTTTTTATTCTGCCATTTTATCAATGAAGCTGGAAATT AACATCATTCACAGATCAACCGTCATCAGACAGACTTTGGAAGAGGACGGAGTTGTCCCA ACTACAGCAGATATTATATATAAGGATGAAACTGCCTCAGAACCACATTTTTTGAGATCT AACGTGGCTATCATTCTGGGAAAAGCATCAGTTATTGGTCTTTTGCTTCTGATCAACCTT TATGTTTTCACAGATAAGTTAAATGCTACAATACTAAACACGGTATATTTTGACTCTACA ATTTACTCGTTACCAAATTTTATCAATTATAAAGATATTGGCAATCTCAGCAATCAAGTG ATCATTTCCGTGTTGCCAAAGCAATATTATACTCCGCTGAAAAAATACCATCAGATCGAA GATTCTGTTCTACTTATCATTGATTCCGTTAGCAATGCTATTCGGGACCAATTTATCAGC AAGTTACTTTTTTTTGCATTTGCAGTTAGTATTTCCATCAATGTCTACTTACTGAATGCT GCAAAAATTCACACAGGATACATGAACTTCCAACCACAATCAAATAAGATCGATGATCTT GTTGTTCAGCAAAAATCGGCAACGATTGAGTTTTCAGAAACTCGAAGTATGCCTGCTTCT TCTGGCCTAGAAACTCCAGTGACCGCGAAAGATATAATTATCTCTGAAGAAATCCAGAAT AACGAATGCGTCTATGCTTTGAGTTCCCAGGACGAGCCTATCCGTCCTTTATCGAATTTA GTGGAACTTATGGAGAAAGAACAATTAAAGAACATGAATAATACTGAGGTTTCGAATCTT GTCGTCAACGGTAAACTGCCATTATATTCCTTAGAGAAAAAATTAGAGGACACAACTCGT GCGGTTTTAGTTAGGAGAAAGGCACTTTCAACTTTGGCTGAATCGCCAATTTTAGTTTCC GAAAAATTGCCCTTCAGAAATTATGATTATGATCGCGTTTTTGGAGCTTGCTGTGAAAAT GTCATCGGCTATATGCCAATACCAGTTGGTGTAATTGGTCCATTAATTATTGATGGAACA TCTTATCACATACCAATGGCAACCACGGAAGGTTGTTTAGTGGCTTCAGCTATGCGTGGT TGCAAAGCCATCAATGCTGGTGGTGGTGCAACAACTGTTTTAACCAAAGATGGTATGACT AGAGGCCCAGTCGTTCGTTTCCCTACTTTAATAAGATCTGGTGCCTGCAAGATATGGTTA GACTCGGAAGAGGGACAAAATTCAATTAAAAAAGCTTTTAATTCTACATCAAGGTTTGCA CGTTTGCAACATATTCAAACCTGTCTAGCAGGCGATTTGCTTTTTATGAGATTTCGGACA ACTACCGGTGACGCAATGGGTATGAACATGATATCGAAAGGTGTCGAATACTCTTTGAAA CAAATGGTAGAAGAATATGGTTGGGAAGATATGGAAGTTGTCTCCGTATCTGGTAACTAT TGTACTGATAAGAAACCTGCCGCAATCAATTGGATTGAAGGTCGTGGTAAAAGTGTCGTA GCTGAAGCTACTATTCCTGGTGATGTCGTAAAAAGTGTTTTAAAGAGCGATGTTTCCGCT TTAGTTGAATTAAATATATCCAAGAACTTGGTTGGATCCGCAATGGCTGGATCTGTTGGT GGTTTCAACGCGCACGCAGCTAATTTGGTCACTGCACTTTTCTTGGCATTAGGCCAAGAT CCTGCGCAGAACGTCGAAAGTTCCAACTGTATAACTTTGATGAAGGAAGTTGATGGTGAT TTAAGGATCTCTGTTTCCATGCCATCTATTGAAGTTGGTACGATTGGCGGGGGTACTGTT CTGGAGCCTCAGGGCGCCATGCTTGATCTTCTCGGCGTTCGTGGTCCTCACCCCACTGAA CCTGGAGCAAATGCTAGGCAATTAGCTAGAATAATCGCGTGTGCTGTCTTGGCTGGTGAA CTGTCTCTGTGCTCCGCACTTGCTGCCGGTCACCTGGTACAAAGCCATATGACTCACAAC CGTAAAACAAACAAAGCCAATGAACTGCCACAACCAAGTAACAAAGGGCCCCCCTGTAAA ACCTCAGCATTATTATAA | ||||||||||||||||||||||||||||
| Protein Properties | |||||||||||||||||||||||||||||
| Pfam Domain Function |
| ||||||||||||||||||||||||||||
| Protein Residues | 1045 | ||||||||||||||||||||||||||||
| Protein Molecular Weight | 115691.0 | ||||||||||||||||||||||||||||
| Protein Theoretical pI | 7.62 | ||||||||||||||||||||||||||||
| Signalling Regions |
| ||||||||||||||||||||||||||||
| Transmembrane Regions |
| ||||||||||||||||||||||||||||
| Protein Sequence | >3-hydroxy-3-methylglutaryl-coenzyme A reductase 2 MSLPLKTIVHLVKPFACTARFSARYPIHVIVVAVLLSAAAYLSVTQSYLNEWKLDSNQYS TYLSIKPDELFEKCTHYYRSPVSDTWKLLSSKEAADIYTPFHYYLSTISFQSKDNSTTLP SLDDVIYSVDHTRYLLSEEPKIPTELVSENGTKWRLRNNSNFILDLHNIYRNMVKQFSNK TSEFDQFDLFIILAAYLTLFYTLCCLFNDMRKIGSKFWLSFSALSNSACALYLSLYTTHS LLKKPASLLSLVIGLPFIVVIIGFKHKVRLAAFSLQKFHRISIDKKITVSNIIYEAMFQE GAYLIRDYLFYISSFIGCAIYARHLPGLVNFCILSTFMLVFDLLLSATFYSAILSMKLEI NIIHRSTVIRQTLEEDGVVPTTADIIYKDETASEPHFLRSNVAIILGKASVIGLLLLINL YVFTDKLNATILNTVYFDSTIYSLPNFINYKDIGNLSNQVIISVLPKQYYTPLKKYHQIE DSVLLIIDSVSNAIRDQFISKLLFFAFAVSISINVYLLNAAKIHTGYMNFQPQSNKIDDL VVQQKSATIEFSETRSMPASSGLETPVTAKDIIISEEIQNNECVYALSSQDEPIRPLSNL VELMEKEQLKNMNNTEVSNLVVNGKLPLYSLEKKLEDTTRAVLVRRKALSTLAESPILVS EKLPFRNYDYDRVFGACCENVIGYMPIPVGVIGPLIIDGTSYHIPMATTEGCLVASAMRG CKAINAGGGATTVLTKDGMTRGPVVRFPTLIRSGACKIWLDSEEGQNSIKKAFNSTSRFA RLQHIQTCLAGDLLFMRFRTTTGDAMGMNMISKGVEYSLKQMVEEYGWEDMEVVSVSGNY CTDKKPAAINWIEGRGKSVVAEATIPGDVVKSVLKSDVSALVELNISKNLVGSAMAGSVG GFNAHAANLVTALFLALGQDPAQNVESSNCITLMKEVDGDLRISVSMPSIEVGTIGGGTV LEPQGAMLDLLGVRGPHPTEPGANARQLARIIACAVLAGELSLCSALAAGHLVQSHMTHN RKTNKANELPQPSNKGPPCKTSALL | ||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||
| External Links |
| ||||||||||||||||||||||||||||
| General Reference |
| ||||||||||||||||||||||||||||