3-hydroxy-3-methylglutaryl-coenzyme A reductase 1 (P12683)
| Identification | |||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Name | 3-hydroxy-3-methylglutaryl-coenzyme A reductase 1 | ||||||||||||||||||||||||||||
| Synonyms |
| ||||||||||||||||||||||||||||
| Gene Name | HMG1 | ||||||||||||||||||||||||||||
| Enzyme Class | |||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||
| General Function | Involved in hydroxymethylglutaryl-CoA reductase (NADPH) activity | ||||||||||||||||||||||||||||
| Specific Function | This transmembrane glycoprotein is involved in the control of cholesterol biosynthesis. It is the rate-limiting enzyme of the sterol biosynthesis | ||||||||||||||||||||||||||||
| Cellular Location | Endoplasmic reticulum membrane; Multi-pass membrane protein | ||||||||||||||||||||||||||||
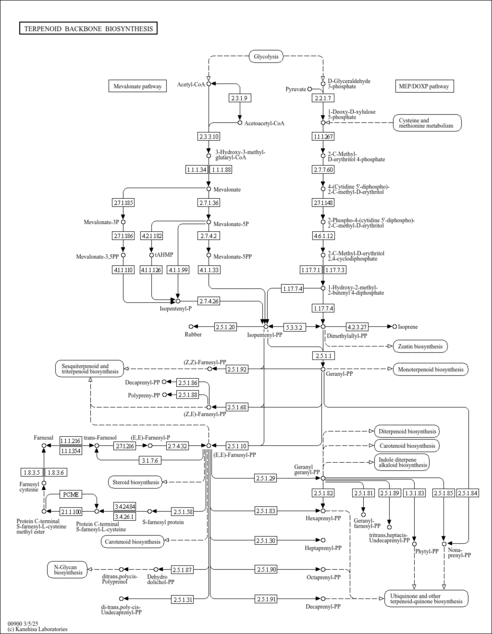
| SMPDB Pathways |
| ||||||||||||||||||||||||||||
| KEGG Pathways |
| ||||||||||||||||||||||||||||
| SMPDB Reactions |
| ||||||||||||||||||||||||||||
| KEGG Reactions |
| ||||||||||||||||||||||||||||
| Metabolites |
| ||||||||||||||||||||||||||||
| GO Classification |
| ||||||||||||||||||||||||||||
| Gene Properties | |||||||||||||||||||||||||||||
| Chromosome Location | chromosome 13 | ||||||||||||||||||||||||||||
| Locus | YML075C | ||||||||||||||||||||||||||||
| Gene Sequence | >3165 bp ATGCCGCCGCTATTCAAGGGACTGAAACAGATGGCAAAGCCAATTGCCTATGTTTCAAGA TTTTCGGCGAAACGACCAATTCATATAATACTTTTTTCTCTAATCATATCCGCATTCGCT TATCTATCCGTCATTCAGTATTACTTCAATGGTTGGCAACTAGATTCAAATAGTGTTTTT GAAACTGCTCCAAATAAAGACTCCAACACTCTATTTCAAGAATGTTCCCATTACTACAGA GATTCCTCTCTAGATGGTTGGGTATCAATCACCGCGCATGAAGCTAGTGAGTTACCAGCC CCACACCATTACTATCTATTAAACCTGAACTTCAATAGTCCTAATGAAACTGACTCCATT CCAGAACTAGCTAACACGGTTTTTGAGAAAGATAATACAAAATATATTCTGCAAGAAGAT CTCAGTGTTTCCAAAGAAATTTCTTCTACTGATGGAACGAAATGGAGGTTAAGAAGTGAC AGAAAAAGTCTTTTCGACGTAAAGACGTTAGCATATTCTCTCTACGATGTATTTTCAGAA AATGTAACCCAAGCAGACCCGTTTGACGTCCTTATTATGGTTACTGCCTACCTAATGATG TTCTACACCATATTCGGCCTCTTCAATGACATGAGGAAGACCGGGTCAAATTTTTGGTTG AGCGCCTCTACAGTGGTCAATTCTGCATCATCACTTTTCTTAGCATTGTATGTCACCCAA TGTATTCTAGGCAAAGAAGTTTCCGCATTAACTCTTTTTGAAGGTTTGCCTTTCATTGTA GTTGTTGTTGGTTTCAAGCACAAAATCAAGATTGCCCAGTATGCCCTGGAGAAATTTGAA AGAGTCGGTTTATCTAAAAGGATTACTACCGATGAAATCGTTTTTGAATCCGTGAGCGAA GAGGGTGGTCGTTTGATTCAAGACCATTTGCTTTGTATTTTTGCCTTTATCGGATGCTCT ATGTATGCTCACCAATTGAAGACTTTGACAAACTTCTGCATATTATCAGCATTTATCCTA ATTTTTGAATTGATTTTAACTCCTACATTTTATTCTGCTATCTTAGCGCTTAGACTGGAA ATGAATGTTATCCACAGATCTACTATTATCAAGCAAACATTAGAAGAAGACGGTGTTGTT CCATCTACAGCAAGAATCATTTCTAAAGCAGAAAAGAAATCCGTATCTTCTTTCTTAAAT CTCAGTGTGGTTGTCATTATCATGAAACTCTCTGTCATACTGTTGTTTGTCTTCATCAAC TTTTATAACTTTGGTGCAAATTGGGTCAATGATGCCTTCAATTCATTGTACTTCGATAAG GAACGTGTTTCTCTACCAGATTTTATTACCTCGAATGCCTCTGAAAACTTTAAAGAGCAA GCTATTGTTAGTGTCACCCCATTATTATATTACAAACCCATTAAGTCCTACCAACGCATT GAGGATATGGTTCTTCTATTGCTTCGTAATGTCAGTGTTGCCATTCGTGATAGGTTCGTC AGTAAATTAGTTCTTTCCGCCTTAGTATGCAGTGCTGTCATCAATGTGTATTTATTGAAT GCTGCTAGAATTCATACCAGTTATACTGCAGACCAATTGGTGAAAACTGAAGTCACCAAG AAGTCTTTTACTGCTCCTGTACAAAAGGCTTCTACACCAGTTTTAACCAATAAAACAGTC ATTTCTGGATCGAAAGTCAAAAGTTTATCATCTGCGCAATCGAGCTCATCAGGACCTTCA TCATCTAGTGAGGAAGATGATTCCCGCGATATTGAAAGCTTGGATAAGAAAATACGTCCT TTAGAAGAATTAGAAGCATTATTAAGTAGTGGAAATACAAAACAATTGAAGAACAAAGAG GTCGCTGCCTTGGTTATTCACGGTAAGTTACCTTTGTACGCTTTGGAGAAAAAATTAGGT GATACTACGAGAGCGGTTGCGGTACGTAGGAAGGCTCTTTCAATTTTGGCAGAAGCTCCT GTATTAGCATCTGATCGTTTACCATATAAAAATTATGACTACGACCGCGTATTTGGCGCT TGTTGTGAAAATGTTATAGGTTACATGCCTTTGCCCGTTGGTGTTATAGGCCCCTTGGTT ATCGATGGTACATCTTATCATATACCAATGGCAACTACAGAGGGTTGTTTGGTAGCTTCT GCCATGCGTGGCTGTAAGGCAATCAATGCTGGCGGTGGTGCAACAACTGTTTTAACTAAG GATGGTATGACAAGAGGCCCAGTAGTCCGTTTCCCAACTTTGAAAAGATCTGGTGCCTGT AAGATATGGTTAGACTCAGAAGAGGGACAAAACGCAATTAAAAAAGCTTTTAACTCTACA TCAAGATTTGCACGTCTGCAACATATTCAAACTTGTCTAGCAGGAGATTTACTCTTCATG AGATTTAGAACAACTACTGGTGACGCAATGGGTATGAATATGATTTCTAAAGGTGTCGAA TACTCATTAAAGCAAATGGTAGAAGAGTATGGCTGGGAAGATATGGAGGTTGTCTCCGTT TCTGGTAACTACTGTACCGACAAAAAACCAGCTGCCATCAACTGGATCGAAGGTCGTGGT AAGAGTGTCGTCGCAGAAGCTACTATTCCTGGTGATGTTGTCAGAAAAGTGTTAAAAAGT GATGTTTCCGCATTGGTTGAGTTGAACATTGCTAAGAATTTGGTTGGATCTGCAATGGCT GGGTCTGTTGGTGGATTTAACGCACATGCAGCTAATTTAGTGACAGCTGTTTTCTTGGCA TTAGGACAAGATCCTGCACAAAATGTTGAAAGTTCCAACTGTATAACATTGATGAAAGAA GTGGACGGTGATTTGAGAATTTCCGTATCCATGCCATCCATCGAAGTAGGTACCATCGGT GGTGGTACTGTTCTAGAACCACAAGGTGCCATGTTGGACTTATTAGGTGTAAGAGGCCCG CATGCTACCGCTCCTGGTACCAACGCACGTCAATTAGCAAGAATAGTTGCCTGTGCCGTC TTGGCAGGTGAATTATCCTTATGTGCTGCCCTAGCAGCCGGCCATTTGGTTCAAAGTCAT ATGACCCACAACAGGAAACCTGCTGAACCAACAAAACCTAACAATTTGGACGCCACTGAT ATAAATCGTTTGAAAGATGGGTCCGTCACCTGCATTAAATCCTAA | ||||||||||||||||||||||||||||
| Protein Properties | |||||||||||||||||||||||||||||
| Pfam Domain Function |
| ||||||||||||||||||||||||||||
| Protein Residues | 1054 | ||||||||||||||||||||||||||||
| Protein Molecular Weight | 115624.0 | ||||||||||||||||||||||||||||
| Protein Theoretical pI | 8.09 | ||||||||||||||||||||||||||||
| Signalling Regions |
| ||||||||||||||||||||||||||||
| Transmembrane Regions |
| ||||||||||||||||||||||||||||
| Protein Sequence | >3-hydroxy-3-methylglutaryl-coenzyme A reductase 1 MPPLFKGLKQMAKPIAYVSRFSAKRPIHIILFSLIISAFAYLSVIQYYFNGWQLDSNSVF ETAPNKDSNTLFQECSHYYRDSSLDGWVSITAHEASELPAPHHYYLLNLNFNSPNETDSI PELANTVFEKDNTKYILQEDLSVSKEISSTDGTKWRLRSDRKSLFDVKTLAYSLYDVFSE NVTQADPFDVLIMVTAYLMMFYTIFGLFNDMRKTGSNFWLSASTVVNSASSLFLALYVTQ CILGKEVSALTLFEGLPFIVVVVGFKHKIKIAQYALEKFERVGLSKRITTDEIVFESVSE EGGRLIQDHLLCIFAFIGCSMYAHQLKTLTNFCILSAFILIFELILTPTFYSAILALRLE MNVIHRSTIIKQTLEEDGVVPSTARIISKAEKKSVSSFLNLSVVVIIMKLSVILLFVFIN FYNFGANWVNDAFNSLYFDKERVSLPDFITSNASENFKEQAIVSVTPLLYYKPIKSYQRI EDMVLLLLRNVSVAIRDRFVSKLVLSALVCSAVINVYLLNAARIHTSYTADQLVKTEVTK KSFTAPVQKASTPVLTNKTVISGSKVKSLSSAQSSSSGPSSSSEEDDSRDIESLDKKIRP LEELEALLSSGNTKQLKNKEVAALVIHGKLPLYALEKKLGDTTRAVAVRRKALSILAEAP VLASDRLPYKNYDYDRVFGACCENVIGYMPLPVGVIGPLVIDGTSYHIPMATTEGCLVAS AMRGCKAINAGGGATTVLTKDGMTRGPVVRFPTLKRSGACKIWLDSEEGQNAIKKAFNST SRFARLQHIQTCLAGDLLFMRFRTTTGDAMGMNMISKGVEYSLKQMVEEYGWEDMEVVSV SGNYCTDKKPAAINWIEGRGKSVVAEATIPGDVVRKVLKSDVSALVELNIAKNLVGSAMA GSVGGFNAHAANLVTAVFLALGQDPAQNVESSNCITLMKEVDGDLRISVSMPSIEVGTIG GGTVLEPQGAMLDLLGVRGPHATAPGTNARQLARIVACAVLAGELSLCAALAAGHLVQSH MTHNRKPAEPTKPNNLDATDINRLKDGSVTCIKS | ||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||
| External Links |
| ||||||||||||||||||||||||||||
| General Reference |
| ||||||||||||||||||||||||||||