Mevalonate kinase (P07277)
| Identification | |||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Name | Mevalonate kinase | ||||||||||||||||||||||||||||||||||||||||||
| Synonyms |
| ||||||||||||||||||||||||||||||||||||||||||
| Gene Name | ERG12 | ||||||||||||||||||||||||||||||||||||||||||
| Enzyme Class | |||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||
| General Function | Involved in ATP binding | ||||||||||||||||||||||||||||||||||||||||||
| Specific Function | RAR (regulation of autonomous replication) is a protein whose activity increases the mitotic stability of plasmids | ||||||||||||||||||||||||||||||||||||||||||
| Cellular Location | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||
| SMPDB Pathways |
| ||||||||||||||||||||||||||||||||||||||||||
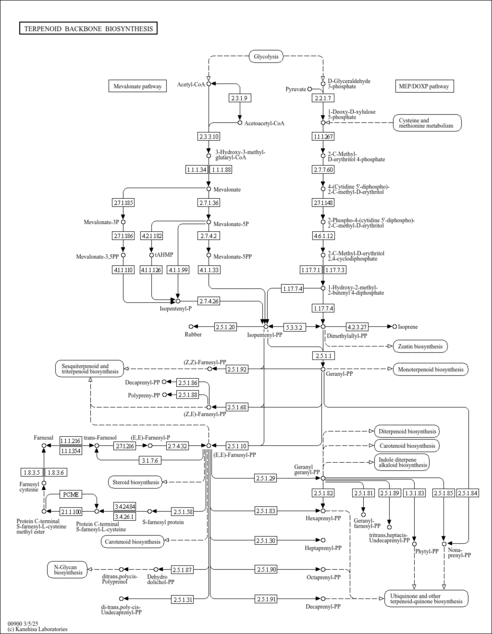
| KEGG Pathways |
| ||||||||||||||||||||||||||||||||||||||||||
| SMPDB Reactions |
| ||||||||||||||||||||||||||||||||||||||||||
| KEGG Reactions | |||||||||||||||||||||||||||||||||||||||||||
| Metabolites |
| ||||||||||||||||||||||||||||||||||||||||||
| GO Classification |
| ||||||||||||||||||||||||||||||||||||||||||
| Gene Properties | |||||||||||||||||||||||||||||||||||||||||||
| Chromosome Location | chromosome 13 | ||||||||||||||||||||||||||||||||||||||||||
| Locus | YMR208W | ||||||||||||||||||||||||||||||||||||||||||
| Gene Sequence | >1332 bp ATGTCATTACCGTTCTTAACTTCTGCACCGGGAAAGGTTATTATTTTTGGTGAACACTCT GCTGTGTACAACAAGCCTGCCGTCGCTGCTAGTGTGTCTGCGTTGAGAACCTACCTGCTA ATAAGCGAGTCATCTGCACCAGATACTATTGAATTGGACTTCCCGGACATTAGCTTTAAT CATAAGTGGTCCATCAATGATTTCAATGCCATCACCGAGGATCAAGTAAACTCCCAAAAA TTGGCCAAGGCTCAACAAGCCACCGATGGCTTGTCTCAGGAACTCGTTAGTCTTTTGGAT CCGTTGTTAGCTCAACTATCCGAATCCTTCCACTACCATGCAGCGTTTTGTTTCCTGTAT ATGTTTGTTTGCCTATGCCCCCATGCCAAGAATATTAAGTTTTCTTTAAAGTCTACTTTA CCCATCGGTGCTGGGTTGGGCTCAAGCGCCTCTATTTCTGTATCACTGGCCTTAGCTATG GCCTACTTGGGGGGGTTAATAGGATCTAATGACTTGGAAAAGCTGTCAGAAAACGATAAG CATATAGTGAATCAATGGGCCTTCATAGGTGAAAAGTGTATTCACGGTACCCCTTCAGGA ATAGATAACGCTGTGGCCACTTATGGTAATGCCCTGCTATTTGAAAAAGACTCACATAAT GGAACAATAAACACAAACAATTTTAAGTTCTTAGATGATTTCCCAGCCATTCCAATGATC CTAACCTATACTAGAATTCCAAGGTCTACAAAAGATCTTGTTGCTCGCGTTCGTGTGTTG GTCACCGAGAAATTTCCTGAAGTTATGAAGCCAATTCTAGATGCCATGGGTGAATGTGCC CTACAAGGCTTAGAGATCATGACTAAGTTAAGTAAATGTAAAGGCACCGATGACGAGGCT GTAGAAACTAATAATGAACTGTATGAACAACTATTGGAATTGATAAGAATAAATCATGGA CTGCTTGTCTCAATCGGTGTTTCTCATCCTGGATTAGAACTTATTAAAAATCTGAGCGAT GATTTGAGAATTGGCTCCACAAAACTTACCGGTGCTGGTGGCGGCGGTTGCTCTTTGACT TTGTTACGAAGAGACATTACTCAAGAGCAAATTGACAGCTTCAAAAAGAAATTGCAAGAT GATTTTAGTTACGAGACATTTGAAACAGACTTGGGTGGGACTGGCTGCTGTTTGTTAAGC GCAAAAAATTTGAATAAAGATCTTAAAATCAAATCCCTAGTATTCCAATTATTTGAAAAT AAAACTACCACAAAGCAACAAATTGACGATCTATTATTGCCAGGAAACACGAATTTACCA TGGACTTCATAA | ||||||||||||||||||||||||||||||||||||||||||
| Protein Properties | |||||||||||||||||||||||||||||||||||||||||||
| Pfam Domain Function |
| ||||||||||||||||||||||||||||||||||||||||||
| Protein Residues | 443 | ||||||||||||||||||||||||||||||||||||||||||
| Protein Molecular Weight | 48458.89844 | ||||||||||||||||||||||||||||||||||||||||||
| Protein Theoretical pI | 5.28 | ||||||||||||||||||||||||||||||||||||||||||
| Signalling Regions |
| ||||||||||||||||||||||||||||||||||||||||||
| Transmembrane Regions |
| ||||||||||||||||||||||||||||||||||||||||||
| Protein Sequence | >Mevalonate kinase MSLPFLTSAPGKVIIFGEHSAVYNKPAVAASVSALRTYLLISESSAPDTIELDFPDISFN HKWSINDFNAITEDQVNSQKLAKAQQATDGLSQELVSLLDPLLAQLSESFHYHAAFCFLY MFVCLCPHAKNIKFSLKSTLPIGAGLGSSASISVSLALAMAYLGGLIGSNDLEKLSENDK HIVNQWAFIGEKCIHGTPSGIDNAVATYGNALLFEKDSHNGTINTNNFKFLDDFPAIPMI LTYTRIPRSTKDLVARVRVLVTEKFPEVMKPILDAMGECALQGLEIMTKLSKCKGTDDEA VETNNELYEQLLELIRINHGLLVSIGVSHPGLELIKNLSDDLRIGSTKLTGAGGGGCSLT LLRRDITQEQIDSFKKKLQDDFSYETFETDLGGTGCCLLSAKNLNKDLKIKSLVFQLFEN KTTTKQQIDDLLLPGNTNLPWTS | ||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||
| External Links |
| ||||||||||||||||||||||||||||||||||||||||||
| General Reference |
| ||||||||||||||||||||||||||||||||||||||||||