| Identification |
|---|
| YMDB ID | YMDB13375 |
|---|
| Name | MG(14:0/0:0/0:0) |
|---|
| Species | Saccharomyces cerevisiae |
|---|
| Strain | Brewer's yeast |
|---|
| Description | MG(14:0/0:0/0:0) belongs to the family of monoradyglycerols, which are glycerolipids lipids containing a common glycerol backbone to which at one fatty acyl group is attached. Their general formula is [R1]OCC(CO[R2])O[R3]. MG(14:0/0:0/0:0) is made up of one tetradecanoyl(R1). |
|---|
| Structure | |
|---|
| Synonyms | - (-)(R)-Glycerol-1-myristate
- 3-Tetradecanoyl-sn-glycerol
- MG (0:0/0:0/14:0)
- sn-3-Tetradecanoylmonoglyceride
- (-)(R)-Glycerol-1-myristic acid
- (2R)-2,3-Dihydroxypropyl tetradecanoic acid
|
|---|
| CAS number | Not Available |
|---|
| Weight | Average: 302.455
Monoisotopic: 302.245709575 |
|---|
| InChI Key | DCBSHORRWZKAKO-MRXNPFEDSA-N |
|---|
| InChI | InChI=1S/C17H34O4/c1-2-3-4-5-6-7-8-9-10-11-12-13-17(20)21-15-16(19)14-18/h16,18-19H,2-15H2,1H3/t16-/m1/s1 |
|---|
| IUPAC Name | (2R)-2,3-dihydroxypropyl tetradecanoate |
|---|
| Traditional IUPAC Name | (2R)-2,3-dihydroxypropyl tetradecanoate |
|---|
| Chemical Formula | C17H34O4 |
|---|
| SMILES | [H][C@@](O)(CO)COC(=O)CCCCCCCCCCCCC |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as 1-monoacylglycerols. These are monoacylglycerols containing a glycerol acylated at the 1-position. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Glycerolipids |
|---|
| Sub Class | Monoradylglycerols |
|---|
| Direct Parent | 1-monoacylglycerols |
|---|
| Alternative Parents | |
|---|
| Substituents | - 1-acyl-sn-glycerol
- Fatty acid ester
- Fatty acyl
- 1,2-diol
- Carboxylic acid ester
- Secondary alcohol
- Carboxylic acid derivative
- Monocarboxylic acid or derivatives
- Primary alcohol
- Hydrocarbon derivative
- Organic oxygen compound
- Carbonyl group
- Alcohol
- Organic oxide
- Organooxygen compound
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Charge | 0 |
|---|
| Melting point | Not Available |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Water Solubility | Not Available | PhysProp | | LogP | Not Available | PhysProp |
|
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations | - Cytoplasm
- Endoplasmic reticulum
|
|---|
| Organoleptic Properties | Not Available |
|---|
| SMPDB Pathways | | Triacylglycerol metabolism TG(10:0/14:0/14:0) | PW007586 |  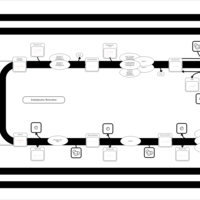 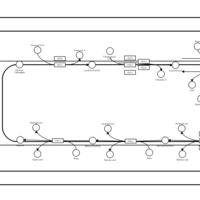 | | Triacylglycerol metabolism TG(10:0/14:0/14:1(9Z)) | PW007590 | 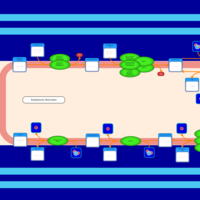 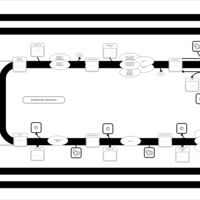  | | Triacylglycerol metabolism TG(10:0/14:0/16:0) | PW007594 | 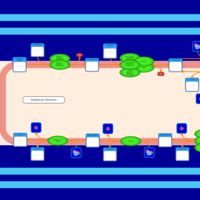   | | Triacylglycerol metabolism TG(10:0/14:0/16:1(9Z)) | PW007600 | 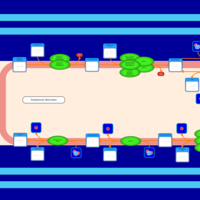  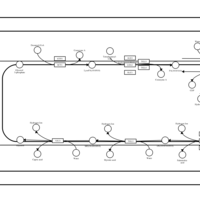 | | Triacylglycerol metabolism TG(10:0/14:0/18:0) | PW007610 |  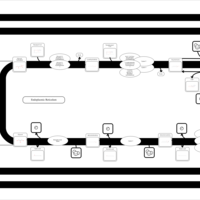 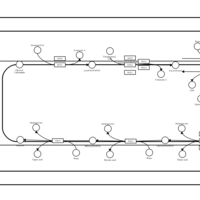 |
|
|---|
| KEGG Pathways | Not Available |
|---|
| SMPDB Reactions | Not Available |
|---|
| KEGG Reactions | Not Available |
|---|
| Concentrations |
|---|
| Intracellular Concentrations | Not Available |
|---|
| Extracellular Concentrations | Not Available |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-053s-9003000000-f85d7c552dc1a4c34131 | JSpectraViewer |
|
|---|
| References |
|---|
| References: | - Rattray JB, Schibeci A, Kidby DK. (1975). "Lipids of yeasts." Bacteriol Rev. 1975 Sep;39(3):197-231.240350
|
|---|
| Synthesis Reference: | Not Available |
|---|
| External Links: | | Resource | Link |
|---|
| CHEBI ID | 75537 | | HMDB ID | Not Available | | Pubchem Compound ID | 71598539 | | Kegg ID | Not Available | | ChemSpider ID | Not Available | | FOODB ID | Not Available | | Wikipedia ID | Not Available | | BioCyc ID | Not Available |
|
|---|
