| Identification |
|---|
| YMDB ID | YMDB01169 |
|---|
| Name | PA(18:1(9Z)/18:1(9Z)) |
|---|
| Species | Saccharomyces cerevisiae |
|---|
| Strain | Brewer's yeast |
|---|
| Description | PA(18:1(9Z)/18:1(9Z))is a phosphatidic acid. It is a glycerophospholipid in which a phosphate moiety occupies a glycerol substitution site. As is the case with diacylglycerols, phosphatidic acids can have many different combinations of fatty acids of varying lengths and saturation attached at the C-1 and C-2 positions. Fatty acids containing 16, 18 and 20 carbons are the most common. PA(18:1(9Z)/18:1(9Z)), in particular, consists of two 9Z-octadecenoyl chain at positions C-1 and C2. The oleic acid moiety is derived from vegetable oils, especially olive and canola oil, while the oleic acid moiety is derived from vegetable oils, especially olive and canola oil. Phosphatidic acids are quite rare but are extremely important as intermediates in the biosynthesis of triacylglycerols and phospholipids. |
|---|
| Structure | |
|---|
| Synonyms | - 1,2-di(9Z-octadecenoyl)-rac-phosphatidic acid
- 1,2-Diacyl-sn-glycerol 3-phosphate
- 1,2-dioleoyl-rac-glycero-3-phosphate
- 1,2-Dioleoyl-sn-glycero-3-phosphate
- 1,2-dioleoylglycerol-3-phosphate
- 3-sn-Phosphatidate
- Dioleoylphosphatidic acid
- DOPA
- PA(18:1(9Z)/18:1(9Z))[U]
- PA(18:1/18:1)
- PA(18:1/18:1)[U]
- PA(18:1n9/18:1n9)
- PA(18:1w9/18:1w9)
- PA(36:2)
- Phosphatidate
- Phosphatidic acid
- Phosphatidic acid(18:1/18:1)
- Phosphatidic acid(18:1n9/18:1n9)
- Phosphatidic acid(18:1w9/18:1w9)
- Phosphatidic acid(36:2)
- 1,2-Di-(9Z-octadecenoyl)-sn-glycero-3-phosphate
- 1-18:1-2-18:1-Phosphatidic acid
- PA(18:1OMEGA9/18:1OMEGA9)
- Phosphatidic acid(18:1omega9/18:1omega9)
- 1,2-Di-(9Z-octadecenoyl)-sn-glycero-3-phosphoric acid
- 1-18:1-2-18:1-Phosphatidate
- Phosphatidate(18:1/18:1)
- Phosphatidate(18:1N9/18:1N9)
- Phosphatidate(18:1OMEGA9/18:1OMEGA9)
- Phosphatidate(36:2)
|
|---|
| CAS number | 14268-17-8 |
|---|
| Weight | Average: 700.979
Monoisotopic: 700.504306309 |
|---|
| InChI Key | MHUWZNTUIIFHAS-DSSVUWSHSA-N |
|---|
| InChI | InChI=1S/C39H73O8P/c1-3-5-7-9-11-13-15-17-19-21-23-25-27-29-31-33-38(40)45-35-37(36-46-48(42,43)44)47-39(41)34-32-30-28-26-24-22-20-18-16-14-12-10-8-6-4-2/h17-20,37H,3-16,21-36H2,1-2H3,(H2,42,43,44)/b19-17-,20-18-/t37-/m1/s1 |
|---|
| IUPAC Name | [(2R)-2,3-bis[(9Z)-octadec-9-enoyloxy]propoxy]phosphonic acid |
|---|
| Traditional IUPAC Name | (2R)-2,3-bis[(9Z)-octadec-9-enoyloxy]propoxyphosphonic acid |
|---|
| Chemical Formula | C39H73O8P |
|---|
| SMILES | [H][C@@](COC(=O)CCCCCCC\C=C/CCCCCCCC)(COP(O)(O)=O)OC(=O)CCCCCCC\C=C/CCCCCCCC |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as 1,2-diacylglycerol-3-phosphates. These are glycerol-3-phosphates in which the glycerol moiety is bonded to two aliphatic chains through ester linkages. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Glycerophospholipids |
|---|
| Sub Class | Glycerophosphates |
|---|
| Direct Parent | 1,2-diacylglycerol-3-phosphates |
|---|
| Alternative Parents | |
|---|
| Substituents | - 1,2-diacylglycerol-3-phosphate
- Fatty acid ester
- Monoalkyl phosphate
- Dicarboxylic acid or derivatives
- Fatty acyl
- Alkyl phosphate
- Phosphoric acid ester
- Organic phosphoric acid derivative
- Carboxylic acid ester
- Carboxylic acid derivative
- Hydrocarbon derivative
- Organic oxide
- Organic oxygen compound
- Carbonyl group
- Organooxygen compound
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Charge | 0 |
|---|
| Melting point | Not Available |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Water Solubility | Not Available | PhysProp | | LogP | Not Available | PhysProp |
|
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations | Not Available |
|---|
| Organoleptic Properties | Not Available |
|---|
| SMPDB Pathways | | Cardiolipin Biosynthesis CL(18:1(9Z)/18:1(9Z)/18:1(9Z)/18:1(11Z)) | PW007898 | 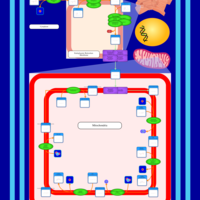 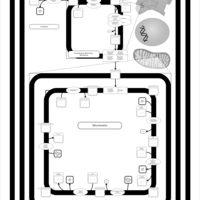 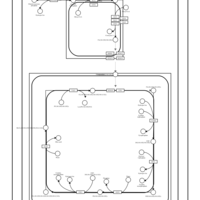 | | Glycerol metabolism | PW002407 | 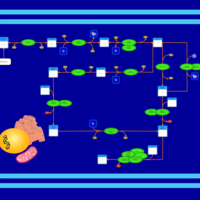 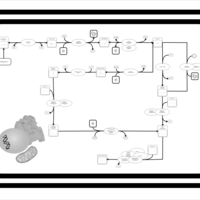 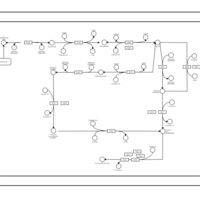 | | Glycerophospholipid metabolism | PW002493 | 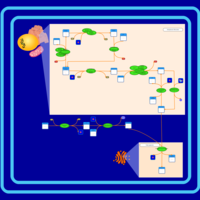 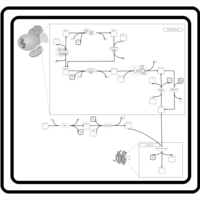 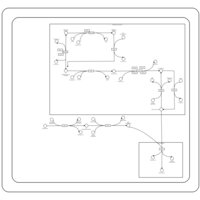 |
|
|---|
| KEGG Pathways | |
|---|
| SMPDB Reactions | Not Available |
|---|
| KEGG Reactions | Not Available |
|---|
| Concentrations |
|---|
| Intracellular Concentrations | Not Available |
|---|
| Extracellular Concentrations | Not Available |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0gb9-1161905400-772a04939f580ad71d3a | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-014i-2393525000-2715e13ba0037dd569f5 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00y0-0196143000-d804e12c01cbda475d7f | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-01sj-4090403000-66c0da756ade4dd045cd | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9050100000-bada02a9f3dfb4726aec | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-60c4b63129e42338dbf2 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-0000009000-55a7c1167cbe0f25b0be | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00l2-1160709000-29509e1741f42c75d166 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-001i-1190301000-37df9990afdea06a6940 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0f89-0000009500-c84c90ec7d2e858c69f3 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0udi-0000007900-6386210cf4e9425daf33 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0uxr-0000509100-163cface94c4f3099804 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00di-0000000900-af2da6ae565960d02a74 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00i0-0000009900-7e41e59856bf50ddbb3c | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-05i3-0000904600-65f32117a29942491df4 | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer |
|
|---|
| References |
|---|
| References: | - Ejsing, C. S., Sampaio, J. L., Surendranath, V., Duchoslav, E., Ekroos, K., Klemm, R. W., Simons, K., Shevchenko, A. (2009). "Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry." Proc Natl Acad Sci U S A 106:2136-2141.19174513
|
|---|
| Synthesis Reference: | Hermetter, A.; Paltauf, F.; Hauser, H. Synthesis of diacyl and alkylacyl glycerophosphoserines. Chemistry and Physics of Lipids (1982), 30(1), 35-45. |
|---|
| External Links: | |
|---|


