| Identification |
|---|
| YMDB ID | YMDB01126 |
|---|
| Name | DG(14:1(9Z)/16:1(9Z)/0:0) |
|---|
| Species | Saccharomyces cerevisiae |
|---|
| Strain | Brewer's yeast |
|---|
| Description | DG(14:1(9Z)/16:1(9Z)/0:0) belongs to the family of Diacylglycerols. These are glycerolipids lipids containing a common glycerol backbone to which at least one fatty acyl group is esterified. DG(14:1(9Z)/16:1(9Z)/0:0) is also a substrate of diacylglycerol kinase. It is involved in the phospholipid metabolic pathway. |
|---|
| Structure | |
|---|
| Synonyms | - 1-myristoleoyl-2-palmitoleoyl-sn-glycerol
- DAG(14:1/16:1)
- DAG(14:1n5/16:1n7)
- DAG(14:1w5/16:1w7)
- DAG(30:2)
- DG(14:1/16:1)
- DG(14:1n5/16:1n7)
- DG(14:1w5/16:1w7)
- DG(30:2)
- Diacylglycerol
- Diacylglycerol(14:1/16:1)
- Diacylglycerol(14:1n5/16:1n7)
- Diacylglycerol(14:1w5/16:1w7)
- Diacylglycerol(30:2)
- Diglyceride
|
|---|
| CAS number | 3738-74-7 |
|---|
| Weight | Average: 536.838
Monoisotopic: 536.444075032 |
|---|
| InChI Key | MMXVUIXFSQFCJO-XCVQYDNVSA-N |
|---|
| InChI | InChI=1S/C33H60O5/c1-3-5-7-9-11-13-15-16-18-20-22-24-26-28-33(36)38-31(29-34)30-37-32(35)27-25-23-21-19-17-14-12-10-8-6-4-2/h10,12-13,15,31,34H,3-9,11,14,16-30H2,1-2H3/b12-10-,15-13-/t31-/m1/s1 |
|---|
| IUPAC Name | (2R)-1-hydroxy-3-[(9Z)-tetradec-9-enoyloxy]propan-2-yl (9Z)-hexadec-9-enoate |
|---|
| Traditional IUPAC Name | (2R)-1-hydroxy-3-[(9Z)-tetradec-9-enoyloxy]propan-2-yl (9Z)-hexadec-9-enoate |
|---|
| Chemical Formula | C33H60O5 |
|---|
| SMILES | [H][C@@](CO)(COC(=O)CCCCCCC\C=C/CCCC)OC(=O)CCCCCCC\C=C/CCCCCC |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as 1,2-diacylglycerols. These are diacylglycerols containing a glycerol acylated at positions 1 and 2. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Glycerolipids |
|---|
| Sub Class | Diradylglycerols |
|---|
| Direct Parent | 1,2-diacylglycerols |
|---|
| Alternative Parents | |
|---|
| Substituents | - 1,2-acyl-sn-glycerol
- Fatty acid ester
- Fatty acyl
- Dicarboxylic acid or derivatives
- Carboxylic acid ester
- Carboxylic acid derivative
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Primary alcohol
- Organooxygen compound
- Carbonyl group
- Alcohol
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Charge | 0 |
|---|
| Melting point | Not Available |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Water Solubility | Not Available | PhysProp | | LogP | Not Available | PhysProp |
|
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations | Not Available |
|---|
| Organoleptic Properties | Not Available |
|---|
| SMPDB Pathways | | Triacylglycerol metabolism TG(14:1(9Z)/15:0/16:1(9Z)) | PW007695 | 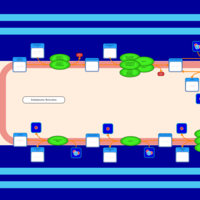 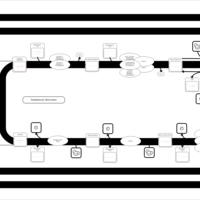 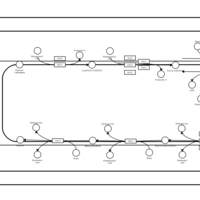 | | Triacylglycerol metabolism TG(14:1(9Z)/16:0/16:1(9Z)) | PW007737 | 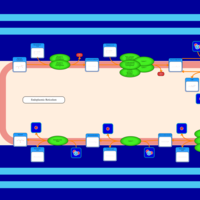 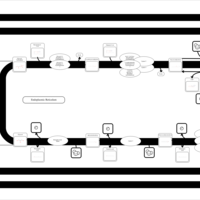  |
|
|---|
| KEGG Pathways | Not Available |
|---|
| SMPDB Reactions | Not Available |
|---|
| KEGG Reactions | Not Available |
|---|
| Concentrations |
|---|
| Intracellular Concentrations | Not Available |
|---|
| Extracellular Concentrations | Not Available |
|---|
| Spectra |
|---|
| Spectra | |
|---|
| References |
|---|
| References: | - Ejsing, C. S., Sampaio, J. L., Surendranath, V., Duchoslav, E., Ekroos, K., Klemm, R. W., Simons, K., Shevchenko, A. (2009). "Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry." Proc Natl Acad Sci U S A 106:2136-2141.19174513
|
|---|
| Synthesis Reference: | Not Available |
|---|
| External Links: | | Resource | Link |
|---|
| CHEBI ID | Not Available | | HMDB ID | HMDB07041 | | Pubchem Compound ID | 1324 | | Kegg ID | C00165 | | ChemSpider ID | 24765875 | | FOODB ID | Not Available | | Wikipedia ID | Not Available | | BioCyc ID | Not Available |
|
|---|
