| Identification |
|---|
| YMDB ID | YMDB00972 |
|---|
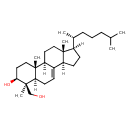
| Name | 4beta-(hydroxymethyl)-4alpha-methyl-5alpha-cholest-7-en-3beta-ol |
|---|
| Species | Saccharomyces cerevisiae |
|---|
| Strain | Baker's yeast |
|---|
| Description | 4beta-hydroxymethyl-4alpha-methyl-5alpha-cholest-7-en-3beta-ol, also known as 4a-methyl-4b-hydroxymethyl-5a-cholest-7-en-3b-ol, belongs to the class of organic compounds known as cholesterols and derivatives. Cholesterols and derivatives are compounds containing a 3-hydroxylated cholestane core. Thus, 4beta-hydroxymethyl-4alpha-methyl-5alpha-cholest-7-en-3beta-ol is considered to be a sterol. Based on a literature review very few articles have been published on 4beta-hydroxymethyl-4alpha-methyl-5alpha-cholest-7-en-3beta-ol. |
|---|
| Structure | |
|---|
| Synonyms | - 4alpha-methyl-4beta-hydroxymethyl-5alpha-cholest-7-en-3beta-ol
- 4beta-Hydroxymethyl-4alpha-methyl-5alpha-cholest-7-en-3beta-ol
- 4a-Methyl-4b-hydroxymethyl-5a-cholest-7-en-3b-ol
- 4Α-methyl-4β-hydroxymethyl-5α-cholest-7-en-3β-ol
- 4b-Hydroxymethyl-4a-methyl-5a-cholest-7-en-3b-ol
- 4Β-hydroxymethyl-4α-methyl-5α-cholest-7-en-3β-ol
- 4b-(Hydroxymethyl)-4a-methyl-5a-cholest-7-en-3b-ol
- 4Β-(hydroxymethyl)-4α-methyl-5α-cholest-7-en-3β-ol
|
|---|
| CAS number | Not Available |
|---|
| Weight | Average: 430.7061
Monoisotopic: 430.381080844 |
|---|
| InChI Key | DWEXIFLNCXYYAA-QQHSWTODSA-N |
|---|
| InChI | InChI=1S/C29H50O2/c1-19(2)8-7-9-20(3)22-11-12-23-21-10-13-25-28(5,24(21)14-16-27(22,23)4)17-15-26(31)29(25,6)18-30/h10,19-20,22-26,30-31H,7-9,11-18H2,1-6H3/t20-,22-,23+,24+,25-,26+,27-,28-,29-/m1/s1 |
|---|
| IUPAC Name | (1R,2R,5S,6S,7R,11R,14R,15R)-6-(hydroxymethyl)-2,6,15-trimethyl-14-[(2R)-6-methylheptan-2-yl]tetracyclo[8.7.0.0²,⁷.0¹¹,¹⁵]heptadec-9-en-5-ol |
|---|
| Traditional IUPAC Name | (1R,2R,5S,6S,7R,11R,14R,15R)-6-(hydroxymethyl)-2,6,15-trimethyl-14-[(2R)-6-methylheptan-2-yl]tetracyclo[8.7.0.0²,⁷.0¹¹,¹⁵]heptadec-9-en-5-ol |
|---|
| Chemical Formula | C29H50O2 |
|---|
| SMILES | [H][C@@]1(CC[C@@]2([H])C3=CC[C@@]4([H])[C@@](C)(CO)[C@@H](O)CC[C@]4(C)[C@@]3([H])CC[C@]12C)[C@H](C)CCCC(C)C |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as cholesterols and derivatives. Cholesterols and derivatives are compounds containing a 3-hydroxylated cholestane core. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Steroids and steroid derivatives |
|---|
| Sub Class | Cholestane steroids |
|---|
| Direct Parent | Cholesterols and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Cholesterol
- Cholesterol-skeleton
- Diterpenoid
- Hydroxysteroid
- 3-hydroxy-delta-7-steroid
- 3-hydroxysteroid
- 3-beta-hydroxysteroid
- Delta-7-steroid
- Cyclic alcohol
- Secondary alcohol
- Primary alcohol
- Organooxygen compound
- Hydrocarbon derivative
- Organic oxygen compound
- Alcohol
- Aliphatic homopolycyclic compound
|
|---|
| Molecular Framework | Aliphatic homopolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Charge | 0 |
|---|
| Melting point | Not Available |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Water Solubility | Not Available | PhysProp | | LogP | Not Available | PhysProp |
|
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations | Not Available |
|---|
| Organoleptic Properties | Not Available |
|---|
| SMPDB Pathways | Not Available |
|---|
| KEGG Pathways | Not Available |
|---|
| SMPDB Reactions | Not Available |
|---|
| KEGG Reactions | Not Available |
|---|
| Concentrations |
|---|
| Intracellular Concentrations | Not Available |
|---|
| Extracellular Concentrations | Not Available |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03e9-0003900000-5251a733d657b50fbd34 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03dj-4139600000-3ea41936e74c41e89115 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0aor-7439100000-15d4b9cac35fedbf23be | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-0001900000-a7a2279b03ba73f5b3e8 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-01u1-0007900000-32e4eaa23d92762230b3 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-001i-1009000000-0822409434a554377d03 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001i-0003900000-544c838c32d262777725 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-02ml-5519300000-22c6a491cf47078b8757 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-052b-9201000000-7af73aca9136d2958ebd | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-0000900000-d3ddef81c134412455e1 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-0000900000-d3ddef81c134412455e1 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-0000900000-91c84e6010c534116b28 | JSpectraViewer | | MS | Mass Spectrum (Electron Ionization) | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer |
|
|---|
| References |
|---|
| References: | - UniProt Consortium (2011). "Ongoing and future developments at the Universal Protein Resource." Nucleic Acids Res 39:D214-D219.21051339
- Scheer, M., Grote, A., Chang, A., Schomburg, I., Munaretto, C., Rother, M., Sohngen, C., Stelzer, M., Thiele, J., Schomburg, D. (2011). "BRENDA, the enzyme information system in 2011." Nucleic Acids Res 39:D670-D676.21062828
- Nose, H., Miyara, T., Kushida, N., Hoshiko, S. (2002). "Isolation of temperature-sensitive Saccharomyces cerevisiae with a mutation in erg25 for C-4 sterol methyl oxidase." J Antibiot (Tokyo) 55:962-968.12546417
- Darnet, S., Rahier, A. (2003). "Enzymological properties of sterol-C4-methyl-oxidase of yeast sterol biosynthesis." Biochim Biophys Acta 1633:106-117.12880870
|
|---|
| Synthesis Reference: | Not Available |
|---|
| External Links: | |
|---|