| Identification |
|---|
| YMDB ID | YMDB00872 |
|---|
| Name | (S)-3-hydroxy-3-methylglutaryl-CoA |
|---|
| Species | Saccharomyces cerevisiae |
|---|
| Strain | Baker's yeast |
|---|
| Description | 3-Hydroxy-3-methylglutaryl-CoA, also known as hydroxymethylglutaroyl coenzyme A or HMG-CoA, belongs to the class of organic compounds known as (s)-3-hydroxy-3-alkylglutaryl coas. These are 3-hydroxy-3-alkylglutaryl-CoAs where the 3-hydroxy-3-alkylglutaryl component has (S)-configuration. Thus, 3-hydroxy-3-methylglutaryl-CoA is considered to be a fatty ester lipid molecule. 3-Hydroxy-3-methylglutaryl-CoA is a very hydrophobic molecule, practically insoluble (in water), and relatively neutral. |
|---|
| Structure | |
|---|
| Synonyms | - (S)-3-Hydroxy-3-methylglutaryl-CoA
- (S)-3-hydroxy-3-methylglutaryl-Coenzyme A
- 3-hydroxy-3-methyl-Glutaryl-CoA
- 3-hydroxy-3-methyl-Glutaryl-Coenzyme A
- 3-hydroxy-3-methylglutaryl-coa
- 3-Hydroxy-3-methylglutaryl-coenzyme A
- hmg-coa
- HMG-Coenzyme A
- hydroxymethylglutaroyl coenzyme a
- Hydroxymethylglutaryl-CoA
- Hydroxymethylglutaryl-Coenzyme A
- S-(Hydrogen 3-hydroxy-3-methylglutaryl)coenzyme A
- S-(Hydrogen 3-hydroxy-3-methylpentanedioate
- S-(Hydrogen 3-hydroxy-3-methylpentanedioate) coenzyme A
- S-(Hydrogen 3-hydroxy-3-methylpentanedioic acid
- (3S)-3-Hydroxy-3-methylglutaryl-coenzyme A
- (3S)-3-Hydroxy-3-methylglutaryl-CoA
- (S)-3-Hydroxy-3-methylglutaryl coenzyme A
- 3-Hydroxy-3-methylglutaryl CoA
- 3-Hydroxy-3-methylglutaryl coenzyme A
- Hydroxymethylglutaryl CoA
- Hydroxymethylglutaryl coenzyme A
- beta-Hydroxy-beta-methylglutaryl CoA
- beta-Hydroxy-beta-methylglutaryl-CoA
- beta-Hydroxy-beta-methylglutaryl-coenzyme A
- Β-hydroxy-β-methylglutaryl CoA
- Β-hydroxy-β-methylglutaryl-CoA
- Β-hydroxy-β-methylglutaryl-coenzyme A
|
|---|
| CAS number | 1553-55-5 |
|---|
| Weight | Average: 911.659
Monoisotopic: 911.157467109 |
|---|
| InChI Key | CABVTRNMFUVUDM-VRHQGPGLSA-N |
|---|
| InChI | InChI=1S/C27H44N7O20P3S/c1-26(2,21(40)24(41)30-5-4-15(35)29-6-7-58-17(38)9-27(3,42)8-16(36)37)11-51-57(48,49)54-56(46,47)50-10-14-20(53-55(43,44)45)19(39)25(52-14)34-13-33-18-22(28)31-12-32-23(18)34/h12-14,19-21,25,39-40,42H,4-11H2,1-3H3,(H,29,35)(H,30,41)(H,36,37)(H,46,47)(H,48,49)(H2,28,31,32)(H2,43,44,45)/t14-,19-,20-,21+,25-,27+/m1/s1 |
|---|
| IUPAC Name | (3S)-5-[(2-{3-[(2R)-3-[({[({[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)oxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy](hydroxy)phosphoryl}oxy)methyl]-2-hydroxy-3-methylbutanamido]propanamido}ethyl)sulfanyl]-3-hydroxy-3-methyl-5-oxopentanoic acid |
|---|
| Traditional IUPAC Name | HMG-CoA |
|---|
| Chemical Formula | C27H44N7O20P3S |
|---|
| SMILES | C[C@](O)(CC(O)=O)CC(=O)SCCNC(=O)CCNC(=O)[C@H](O)C(C)(C)COP(O)(=O)OP(O)(=O)OC[C@H]1O[C@H]([C@H](O)[C@@H]1OP(O)(O)=O)N1C=NC2=C1N=CN=C2N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as (s)-3-hydroxy-3-alkylglutaryl coas. These are 3-hydroxy-3-alkylglutaryl-CoAs where the 3-hydroxy-3-alkylglutaryl component has (S)-configuration. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Fatty Acyls |
|---|
| Sub Class | Fatty acyl thioesters |
|---|
| Direct Parent | (S)-3-hydroxy-3-alkylglutaryl CoAs |
|---|
| Alternative Parents | |
|---|
| Substituents | - Coenzyme a or derivatives
- Purine ribonucleoside 3',5'-bisphosphate
- Purine ribonucleoside bisphosphate
- Purine ribonucleoside diphosphate
- Ribonucleoside 3'-phosphate
- Pentose phosphate
- Pentose-5-phosphate
- Beta amino acid or derivatives
- Glycosyl compound
- N-glycosyl compound
- 6-aminopurine
- Monosaccharide phosphate
- Organic pyrophosphate
- Pentose monosaccharide
- Imidazopyrimidine
- Purine
- Monoalkyl phosphate
- Aminopyrimidine
- Alkyl phosphate
- Imidolactam
- N-acyl-amine
- N-substituted imidazole
- Organic phosphoric acid derivative
- Monosaccharide
- Fatty amide
- Pyrimidine
- Phosphoric acid ester
- Tetrahydrofuran
- Tertiary alcohol
- Imidazole
- Heteroaromatic compound
- Azole
- Secondary alcohol
- Thiocarboxylic acid ester
- Amino acid
- Carboxamide group
- Amino acid or derivatives
- Carbothioic s-ester
- Secondary carboxylic acid amide
- Organoheterocyclic compound
- Sulfenyl compound
- Thiocarboxylic acid or derivatives
- Azacycle
- Oxacycle
- Carboxylic acid derivative
- Carboxylic acid
- Monocarboxylic acid or derivatives
- Organic oxygen compound
- Primary amine
- Hydrocarbon derivative
- Carbonyl group
- Organosulfur compound
- Organopnictogen compound
- Organic oxide
- Organooxygen compound
- Organonitrogen compound
- Organic nitrogen compound
- Alcohol
- Amine
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Charge | 0 |
|---|
| Melting point | Not Available |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Water Solubility | Not Available | PhysProp | | LogP | Not Available | PhysProp |
|
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations | Not Available |
|---|
| Organoleptic Properties | Not Available |
|---|
| SMPDB Pathways | | Cholesterol biosynthesis and metabolism CE(10:0) | PW002545 | 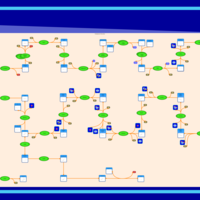 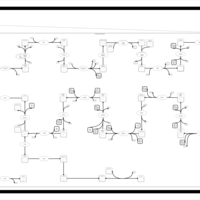 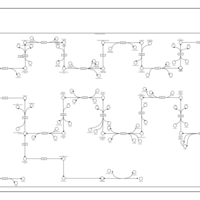 | | Cholesterol biosynthesis and metabolism CE(12:0) | PW002548 | 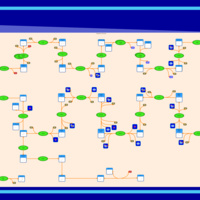   | | Cholesterol biosynthesis and metabolism CE(14:0) | PW002544 |    | | Cholesterol biosynthesis and metabolism CE(16:0) | PW002550 |    | | Cholesterol biosynthesis and metabolism CE(18:0) | PW002551 |    |
|
|---|
| KEGG Pathways | | Butanoate metabolism | ec00650 |  | | Steroid biosynthesis | ec00100 |  | | Synthesis and degradation of ketone bodies | ec00072 | 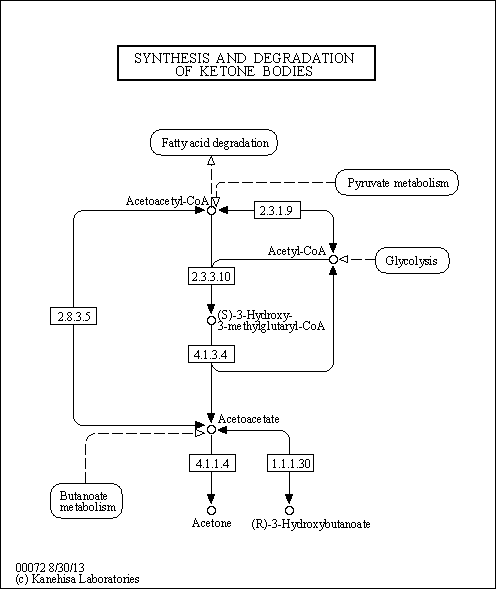 | | Terpenoid backbone biosynthesis | ec00900 |  | | Valine, leucine and isoleucine degradation | ec00280 |  |
|
|---|
| SMPDB Reactions | Not Available |
|---|
| KEGG Reactions | Not Available |
|---|
| Concentrations |
|---|
| Intracellular Concentrations | Not Available |
|---|
| Extracellular Concentrations | Not Available |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000l-0891000250-052d51ef28faa9ab5b73 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-0971100000-dc16942c0a4531c8cb60 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-1950000000-12dcb50a3caaf20a9aa5 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00o4-4901031462-49fbf273ff8df18347bb | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-001i-3900110010-0be1f0212b5d814ea24c | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-057i-6900100000-fffa063c59d4fcd54e7a | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03dm-0000000094-b818de142848216a6af3 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4u-9500001521-8d0d8c41c4c284cc696f | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004m-9202503430-001456d7b42e54584702 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-0000000169-47d0ed82d5acca38b9e0 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-002r-1500000391-9d63a322b718612b8587 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4i-0011900000-766a56d8ca4793434590 | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer |
|
|---|
| References |
|---|
| References: | - Scheer, M., Grote, A., Chang, A., Schomburg, I., Munaretto, C., Rother, M., Sohngen, C., Stelzer, M., Thiele, J., Schomburg, D. (2011). "BRENDA, the enzyme information system in 2011." Nucleic Acids Res 39:D670-D676.21062828
- Middleton, B. (1972). "The kinetic mechanism of 3-hydroxy-3-methylglutaryl-coenzyme A synthase from baker's yeast." Biochem J 126:35-47.4561618
- Serafin, J., Kreczko, R. (1979). "[Use of transcondylar and metaphyseal osteotomies of the tibia in the treatment of degenerative changes of the knee joint]." Chir Narzadow Ruchu Ortop Pol 44:141-148.456161
|
|---|
| Synthesis Reference: | Not Available |
|---|
| External Links: | |
|---|





