| Identification |
|---|
| YMDB ID | YMDB00844 |
|---|
| Name | PPPi |
|---|
| Species | Saccharomyces cerevisiae |
|---|
| Strain | Baker's yeast |
|---|
| Description | Triphosphate, also known as H5P3O10 or triphosphorsaeure, belongs to the class of inorganic compounds known as non-metal phosphates. These are inorganic non-metallic compounds containing a phosphate as its largest oxoanion. Triphosphate is an extremely strong acidic compound (based on its pKa). Triphosphate exists in all living species, ranging from bacteria to humans. |
|---|
| Structure | |
|---|
| Synonyms | - (phosphate)n
- (Phosphate)n-1
- (Phosphate)n+1
- acide triphosphorique
- Bis(dihydroxidodioxidophosphato)hydroxidooxidophosphorus
- Bis(phosphonooxy)phosphinic acid
- Catena-triphosphoric acid
- DAD
- DCT
- DGT
- Diphosphono hydrogen phosphate
- DTP
- GTP
- Inorganic open chain tripolyphosphate
- Inorganic triphosphate
- Triphosphate
- Triphosphate analogs
- Triphosphoric acid
- Triphosphorsaeure
- Tripolyphosphate
- TTP
- H5P3O10
- Tripolyphosphoric acid
- Catena-triphosphate
- Inorganic triphosphoric acid
- Triphospate
- Sodium triphosphate
- Tetrasodium tripolyphosphate
- Triphosphoric acid, 99TC-labeled CPD
- Triphosphoric acid, pentasodium salt
- Triphosphoric acid, sodium salt
- Potassium triphosphate
- Sodium tripolyphosphate anhydrous
- Triphosphoric acid, pentapotassium salt
- Triphosphoric acid, sodium, potassium salt
- Pentapotassium triphosphate
- Sodium tripolyphosphate
- PPPi
|
|---|
| CAS number | 10380-08-2 |
|---|
| Weight | Average: 257.955
Monoisotopic: 257.909555916 |
|---|
| InChI Key | UNXRWKVEANCORM-UHFFFAOYSA-N |
|---|
| InChI | InChI=1S/H5O10P3/c1-11(2,3)9-13(7,8)10-12(4,5)6/h(H,7,8)(H2,1,2,3)(H2,4,5,6) |
|---|
| IUPAC Name | {[hydroxy(phosphonooxy)phosphoryl]oxy}phosphonic acid |
|---|
| Traditional IUPAC Name | tripolyphosphate |
|---|
| Chemical Formula | H5O10P3 |
|---|
| SMILES | [H]OP(=O)(O[H])OP(=O)(O[H])OP(=O)(O[H])O[H] |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of inorganic compounds known as non-metal phosphates. These are inorganic non-metallic compounds containing a phosphate as its largest oxoanion. |
|---|
| Kingdom | Inorganic compounds |
|---|
| Super Class | Homogeneous non-metal compounds |
|---|
| Class | Non-metal oxoanionic compounds |
|---|
| Sub Class | Non-metal phosphates |
|---|
| Direct Parent | Non-metal phosphates |
|---|
| Alternative Parents | |
|---|
| Substituents | - Non-metal phosphate
- Inorganic oxide
|
|---|
| Molecular Framework | Not Available |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Charge | 0 |
|---|
| Melting point | Not Available |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Water Solubility | Not Available | PhysProp | | LogP | Not Available | PhysProp |
|
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations | Not Available |
|---|
| Organoleptic Properties | Not Available |
|---|
| SMPDB Pathways | | Oxidative phosphorylation | PW002461 | 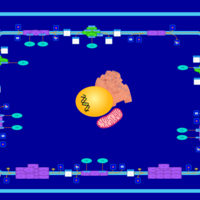 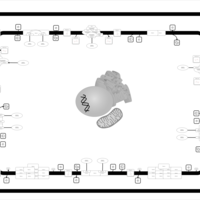 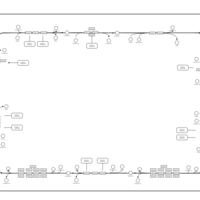 | | purine nucleotides de novo biosynthesis | PW002478 |  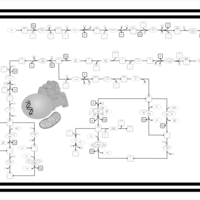 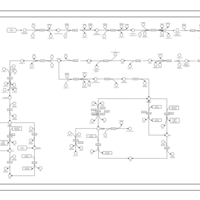 |
|
|---|
| KEGG Pathways | |
|---|
| SMPDB Reactions | Not Available |
|---|
| KEGG Reactions | Not Available |
|---|
| Concentrations |
|---|
| Intracellular Concentrations | Not Available |
|---|
| Extracellular Concentrations | Not Available |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0002-9520000000-e90a8c73374acf7bc122 | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0a6r-2690000000-673654046903650b7152 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03fs-5930000000-5eb4974ca8fed7558024 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-01qa-9500000000-b2404c5ea6e713839384 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0090000000-7ab59c02dffe21b7a604 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-056r-9270000000-61dc72f8bf5e400f3882 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-6a7c993cb33cf4d5fedd | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0a4i-0090000000-db0b792e00f5267b0429 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a6r-4690000000-1c8ad94ae184a8e58b61 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0002-9000000000-9afb7f62828d4245d383 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0390000000-05444cfee24772b7b3b2 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4i-0900000000-2eca580fe4fd0b82d8a6 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-0900000000-2e0c12969f0c2680414e | JSpectraViewer |
|
|---|
| References |
|---|
| References: | - Scheer, M., Grote, A., Chang, A., Schomburg, I., Munaretto, C., Rother, M., Sohngen, C., Stelzer, M., Thiele, J., Schomburg, D. (2011). "BRENDA, the enzyme information system in 2011." Nucleic Acids Res 39:D670-D676.21062828
|
|---|
| Synthesis Reference: | Not Available |
|---|
| External Links: | |
|---|

