| Identification |
|---|
| YMDB ID | YMDB00797 |
|---|
| Name | udp-galactose |
|---|
| Species | Saccharomyces cerevisiae |
|---|
| Strain | Baker's yeast |
|---|
| Description | udp-galactose belongs to the class of organic compounds known as pentose phosphates. These are carbohydrate derivatives containing a pentose substituted by one or more phosphate groups. udp-galactose is an extremely weak basic (essentially neutral) compound (based on its pKa). udp-galactose may be a unique S. cerevisiae (yeast) metabolite. |
|---|
| Structure | |
|---|
| Synonyms | - GALACTOSE-URIDINE-5'-DIPHOSPHATE
- GDU
- Udp galactose
- UDP-a-D-Galactose
- Udp-alpha-d-galactose
- UDP-alpha-delta-Galactose
- udp-d-galactopyranose
- udp-d-galactose
- Udp-delta-galactopyranose
- UDP-delta-galactose
- UDP-Gal
- UDP-galactopyranose
- UDP-galactose
- Udpgal
- UDPgalactose
- UPG
- Uridine 5'-(alpha-D-galactopyranosyl pyrophosphate)
- Uridine 5'-(alpha-delta-galactopyranosyl pyrophosphate)
- uridine 5'-[3-(D-galactopyranosyl) dihydrogen diphosphate]
- Uridine 5'-diphosphate galactose
- Uridine 5'-diphosphogalactose
- Uridine 5'-pyrophosphate a-D-galactopyranosyl ester
- Uridine 5'-pyrophosphate a-D-galactosyl ester
- Uridine 5'-pyrophosphate a-delta-galactopyranosyl ester
- Uridine 5'-pyrophosphate a-delta-galactosyl ester
- Uridine 5'-pyrophosphate alpha-D-galactosyl ester
- Uridine 5'-pyrophosphate alpha-delta-galactosyl ester
- Uridine 5'-pyrophosphate D-galactosyl ester
- Uridine diphosphate galactose
- Uridine diphosphate-D-galactose
- Uridine diphosphate-delta-galactose
- Uridine diphosphate-galactose
- Uridine diphosphogalactose
- Uridine pyrophosphate a-D-galactopyranosyl ester
- Uridine pyrophosphate a-delta-galactopyranosyl ester
- Uridine pyrophosphate alpha-d-galactopyranosyl ester
- Uridine pyrophosphate alpha-delta-galactopyranosyl ester
- Uridine pyrophosphogalactose
- Uridinediphosphate galactose
- Uridinediphosphogalactose
|
|---|
| CAS number | 2956-16-3 |
|---|
| Weight | Average: 566.3018
Monoisotopic: 566.055020376 |
|---|
| InChI Key | USAZACJQJDHAJH-QFIAFIRLSA-N |
|---|
| InChI | InChI=1S/C15H24N2O17P2/c18-2-5-8(20)10(22)12(24)14(32-5)33-36(28,29)34-35(26,27)30-3-6-9(21)11(23)13(31-6)4-1-7(19)17-15(25)16-4/h1,5-6,8-14,18,20-24H,2-3H2,(H,26,27)(H,28,29)(H2,16,17,19,25)/t5-,6-,8+,9-,10+,11-,12-,13+,14+/m1/s1 |
|---|
| IUPAC Name | [({[(2R,3S,4R,5S)-5-(2,6-dioxo-1,2,3,6-tetrahydropyrimidin-4-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy]({[(2S,3R,4S,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy})phosphinic acid |
|---|
| Traditional IUPAC Name | {[(2R,3S,4R,5S)-5-(2,6-dioxo-1,3-dihydropyrimidin-4-yl)-3,4-dihydroxyoxolan-2-yl]methoxy(hydroxy)phosphoryl}oxy[(2S,3R,4S,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxyphosphinic acid |
|---|
| Chemical Formula | C15H24N2O17P2 |
|---|
| SMILES | OC[C@H]1O[C@@H](OP(O)(=O)OP(O)(=O)OC[C@H]2O[C@H]([C@H](O)[C@@H]2O)C2=CC(=O)NC(=O)N2)[C@H](O)[C@@H](O)[C@H]1O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as pentose phosphates. These are carbohydrate derivatives containing a pentose substituted by one or more phosphate groups. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | Pentose phosphates |
|---|
| Alternative Parents | |
|---|
| Substituents | - Pentose-5-phosphate
- Pentose phosphate
- Glycosyl compound
- C-glycosyl compound
- Organic pyrophosphate
- Monosaccharide phosphate
- Monoalkyl phosphate
- Pyrimidone
- Alkyl phosphate
- Pyrimidine
- Phosphoric acid ester
- Oxane
- Organic phosphoric acid derivative
- Hydropyrimidine
- Heteroaromatic compound
- Vinylogous amide
- Tetrahydrofuran
- Urea
- Secondary alcohol
- Lactam
- Oxacycle
- Azacycle
- Organoheterocyclic compound
- Polyol
- Ether
- Dialkyl ether
- Organic nitrogen compound
- Organopnictogen compound
- Organic oxide
- Hydrocarbon derivative
- Primary alcohol
- Organonitrogen compound
- Alcohol
- Aromatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aromatic heteromonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Charge | 0 |
|---|
| Melting point | Not Available |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Water Solubility | Not Available | PhysProp | | LogP | Not Available | PhysProp |
|
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations | Not Available |
|---|
| Organoleptic Properties | Not Available |
|---|
| SMPDB Pathways | | Amino sugar and nucleotide sugar metabolism | PW002413 | 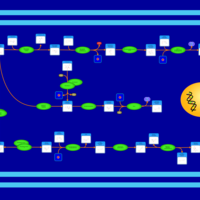 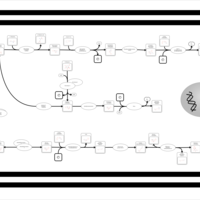 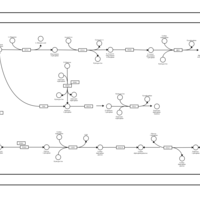 |
|
|---|
| KEGG Pathways | | Amino sugar and nucleotide sugar metabolism | ec00520 |  | | Galactose metabolism | ec00052 |  |
|
|---|
| SMPDB Reactions | Not Available |
|---|
| KEGG Reactions | Not Available |
|---|
| Concentrations |
|---|
| Intracellular Concentrations | Not Available |
|---|
| Extracellular Concentrations | Not Available |
|---|
| Spectra |
|---|
| Spectra | |
|---|
| References |
|---|
| References: | - UniProt Consortium (2011). "Ongoing and future developments at the Universal Protein Resource." Nucleic Acids Res 39:D214-D219.21051339
- Scheer, M., Grote, A., Chang, A., Schomburg, I., Munaretto, C., Rother, M., Sohngen, C., Stelzer, M., Thiele, J., Schomburg, D. (2011). "BRENDA, the enzyme information system in 2011." Nucleic Acids Res 39:D670-D676.21062828
- DOUGLAS, H. C., HAWTHORNE, D. C. (1964). "ENZYMATIC EXPRESSION AND GENETIC LINKAGE OF GENES CONTROLLING GALACTOSE UTILIZATION IN SACCHAROMYCES." Genetics 49:837-844.14158615
|
|---|
| Synthesis Reference: | Not Available |
|---|
| External Links: | |
|---|


