| Identification |
|---|
| YMDB ID | YMDB00749 |
|---|
| Name | (R)-4'-phosphopantothenic acid |
|---|
| Species | Saccharomyces cerevisiae |
|---|
| Strain | Baker's yeast |
|---|
| Description | D-4'-Phosphopantothenate, also known as phosphopantothenic acid, belongs to the class of organic compounds known as beta amino acids and derivatives. These are amino acids having a (-NH2) group attached to the beta carbon atom. D-4'-Phosphopantothenate is an extremely weak basic (essentially neutral) compound (based on its pKa). Within yeast, D-4'-phosphopantothenate participates in a number of enzymatic reactions. In particular, D-4'-phosphopantothenate can be biosynthesized from pantothenic acid; which is mediated by the enzyme pantothenate kinase. In addition, D-4'-phosphopantothenate, cytidine triphosphate, and L-cysteine can be converted into cytidine monophosphate and 4'-phosphopantothenoylcysteine through the action of the enzyme phosphopantothenate-cysteine ligase. In yeast, D-4'-phosphopantothenate is involved in the metabolic pathway called the beta-alanine metabolism pathway. |
|---|
| Structure | |
|---|
| Synonyms | - (r)-4-phosphopantothenate
- (R)-4'-phosphopantothenate
- (R)-N-(2-hydroxy-3,3-dimethyl-1-oxo-4-(phosphonooxy)butyl)-beta-alanine
- 4'-P-Pantothenate
- 4'-phosphopantothenate
- D-4'-Phosphopantothenate
- D-4'-Phosphopantothenic acid
- N-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alanine
- phosphopantothenic acid
- (R)-4'-Phosphopantothenic acid
- (R)-N-(2-Hydroxy-3,3-dimethyl-1-oxo-4-(phosphonooxy)butyl)-b-alanine
- (R)-N-(2-Hydroxy-3,3-dimethyl-1-oxo-4-(phosphonooxy)butyl)-β-alanine
- N-[(2R)-2-Hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-b-alanine
- N-[(2R)-2-Hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-β-alanine
- Phosphopantothenate
- Phosphopantothenic acid, calcium salt (2:1)
- Phosphopantothenic acid, calcium salt, (R)-isomer
- 4'-Phospho-D-pantothenic acid
- 4'-Phosphopantothenic acid
- 4’-phospho-D-pantothenic acid
- 4’-phosphopantothenic acid
- D-Pantothenic acid 4'-phosphate
- D-Pantothenic acid 4’-phosphate
- N-[(2R)-2-Hydroxy-3,3-dimethyl-1-oxo-4-(phosphonooxy)butyl]-beta-alanine
- N-[(2R)-2-Hydroxy-3,3-dimethyl-1-oxo-4-(phosphonooxy)butyl]-β-alanine
- (R)-4'-Phosphonatopantothenate
- (R)-4’-phosphonatopantothenate
- (R)-4’-phosphopantothenate
- 4’-phosphopantothenate
- D-4’-phosphopantothenate
- D-4’-phosphopantothenic acid
|
|---|
| CAS number | Not Available |
|---|
| Weight | Average: 299.2149
Monoisotopic: 299.077003069 |
|---|
| InChI Key | XHFVGHPGDLDEQO-ZETCQYMHSA-N |
|---|
| InChI | InChI=1S/C9H18NO8P/c1-9(2,5-18-19(15,16)17)7(13)8(14)10-4-3-6(11)12/h7,13H,3-5H2,1-2H3,(H,10,14)(H,11,12)(H2,15,16,17)/t7-/m0/s1 |
|---|
| IUPAC Name | 3-[(2R)-2-hydroxy-3-methyl-3-[(phosphonooxy)methyl]butanamido]propanoic acid |
|---|
| Traditional IUPAC Name | phosphopantothenic acid |
|---|
| Chemical Formula | C9H18NO8P |
|---|
| SMILES | CC(C)(COP(O)(O)=O)[C@@H](O)C(=O)NCCC(O)=O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as beta amino acids and derivatives. These are amino acids having a (-NH2) group attached to the beta carbon atom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | Beta amino acids and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Beta amino acid or derivatives
- Monoalkyl phosphate
- Fatty amide
- Monosaccharide
- N-acyl-amine
- Organic phosphoric acid derivative
- Phosphoric acid ester
- Alkyl phosphate
- Fatty acyl
- Secondary alcohol
- Carboxamide group
- Secondary carboxylic acid amide
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Organooxygen compound
- Organonitrogen compound
- Carbonyl group
- Organic nitrogen compound
- Organic oxygen compound
- Alcohol
- Hydrocarbon derivative
- Organic oxide
- Organopnictogen compound
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Charge | 0 |
|---|
| Melting point | Not Available |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Water Solubility | Not Available | PhysProp | | LogP | Not Available | PhysProp |
|
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations | Not Available |
|---|
| Organoleptic Properties | Not Available |
|---|
| SMPDB Pathways | |
|---|
| KEGG Pathways | | Pantothenate and CoA biosynthesis | ec00770 | 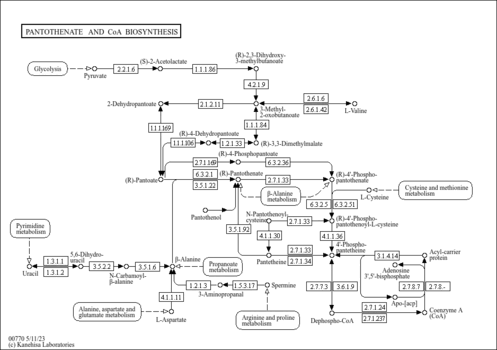 | | beta-Alanine metabolism | ec00410 |  |
|
|---|
| SMPDB Reactions | Not Available |
|---|
| KEGG Reactions | Not Available |
|---|
| Concentrations |
|---|
| Intracellular Concentrations | Not Available |
|---|
| Extracellular Concentrations | Not Available |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-000t-9720000000-41704091b1bf13376017 | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0f79-9141000000-5b0b47ec4bcdae4b9de9 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-9320000000-cfdd61fa71ff49880f5d | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0076-9000000000-c93d1a5c4707410e2d61 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-002b-9270000000-b61365a2ce574c07d431 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9100000000-ead403fde97da638fdc5 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-57aba113e2388577d6a9 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-2296000000-391c82c7a505e9c802ca | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0udj-9241000000-b0f7eb9f3a7d6f4ec655 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0udu-8920000000-0c09af15c101c82e1d13 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000t-0090000000-832ae76b1a16ae3ea75c | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004j-4090000000-f6c91ebb49be49cd09ec | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9200000000-1f482bd5f3b51408f3af | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer |
|
|---|
| References |
|---|
| References: | - Scheer, M., Grote, A., Chang, A., Schomburg, I., Munaretto, C., Rother, M., Sohngen, C., Stelzer, M., Thiele, J., Schomburg, D. (2011). "BRENDA, the enzyme information system in 2011." Nucleic Acids Res 39:D670-D676.21062828
- Herrgard, M. J., Swainston, N., Dobson, P., Dunn, W. B., Arga, K. Y., Arvas, M., Bluthgen, N., Borger, S., Costenoble, R., Heinemann, M., Hucka, M., Le Novere, N., Li, P., Liebermeister, W., Mo, M. L., Oliveira, A. P., Petranovic, D., Pettifer, S., Simeonidis, E., Smallbone, K., Spasic, I., Weichart, D., Brent, R., Broomhead, D. S., Westerhoff, H. V., Kirdar, B., Penttila, M., Klipp, E., Palsson, B. O., Sauer, U., Oliver, S. G., Mendes, P., Nielsen, J., Kell, D. B. (2008). "A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology." Nat Biotechnol 26:1155-1160.18846089
|
|---|
| Synthesis Reference: | Not Available |
|---|
| External Links: | |
|---|


