| Identification |
|---|
| YMDB ID | YMDB00664 |
|---|
| Name | 3'-Dephospho-CoA |
|---|
| Species | Saccharomyces cerevisiae |
|---|
| Strain | Baker's yeast |
|---|
| Description | Dephospho Coenzyme A, also known as dephospho-CoA or dpcoa, belongs to the class of organic compounds known as purine ribonucleoside diphosphates. These are purine ribobucleotides with diphosphate group linked to the ribose moiety. Dephospho Coenzyme A is a strong basic compound (based on its pKa). Dephospho Coenzyme A exists in both E. coli (prokaryote) and yeast (eukaryote). |
|---|
| Structure | |
|---|
| Synonyms | - 3'-dephospho-Coenzyme A
- 3'-O-dephosphono-CoA
- 3'-O-dephosphono-Coenzyme A
- Coenzyme A, 3'-O-dephosphono-
- Dephospho coa
- Dephospho-CoA
- Dephospho-Coenzyme A
- dephosphocoenzyme a
- 3'-Dephospho-CoA
- DpCoA
- 2'-(5''-Triphosphoribosyl)-3'-dephospho-CoA
- 3'-Desphospho-coenzyme A
- Desphospho-CoA
- 3'-Dephosphocoenzyme A
- 3'-O-Dephosphonocoenzyme A
- 3’-Dephospho-CoA
- 3’-Dephospho-coenzyme A
- 3’-Dephosphocoenzyme A
- 3’-O-Dephosphonocoenzyme A
|
|---|
| CAS number | 3633-59-8 |
|---|
| Weight | Average: 687.554
Monoisotopic: 687.148877955 |
|---|
| InChI Key | KDTSHFARGAKYJN-IBOSZNHHSA-N |
|---|
| InChI | InChI=1S/C21H35N7O13P2S/c1-21(2,16(32)19(33)24-4-3-12(29)23-5-6-44)8-39-43(36,37)41-42(34,35)38-7-11-14(30)15(31)20(40-11)28-10-27-13-17(22)25-9-26-18(13)28/h9-11,14-16,20,30-32,44H,3-8H2,1-2H3,(H,23,29)(H,24,33)(H,34,35)(H,36,37)(H2,22,25,26)/t11-,14-,15-,16+,20-/m1/s1 |
|---|
| IUPAC Name | [({[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy][(3R)-3-hydroxy-2,2-dimethyl-3-({2-[(2-sulfanylethyl)carbamoyl]ethyl}carbamoyl)propoxy]phosphinic acid |
|---|
| Traditional IUPAC Name | 3'-dephospho-coa |
|---|
| Chemical Formula | C21H35N7O13P2S |
|---|
| SMILES | CC(C)(COP(O)(=O)OP(O)(=O)OC[C@H]1O[C@H]([C@H](O)[C@@H]1O)N1C=NC2=C1N=CN=C2N)[C@@H](O)C(=O)NCCC(=O)NCCS |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as purine ribonucleoside diphosphates. These are purine ribobucleotides with diphosphate group linked to the ribose moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Nucleosides, nucleotides, and analogues |
|---|
| Class | Purine nucleotides |
|---|
| Sub Class | Purine ribonucleotides |
|---|
| Direct Parent | Purine ribonucleoside diphosphates |
|---|
| Alternative Parents | |
|---|
| Substituents | - Purine ribonucleoside diphosphate
- Purine ribonucleoside monophosphate
- Pentose phosphate
- Pentose-5-phosphate
- Beta amino acid or derivatives
- Glycosyl compound
- N-glycosyl compound
- 6-aminopurine
- Pentose monosaccharide
- Monosaccharide phosphate
- Organic pyrophosphate
- Imidazopyrimidine
- Purine
- Aminopyrimidine
- Monoalkyl phosphate
- N-acyl-amine
- N-substituted imidazole
- Organic phosphoric acid derivative
- Pyrimidine
- Fatty amide
- Imidolactam
- Monosaccharide
- Fatty acyl
- Phosphoric acid ester
- Alkyl phosphate
- Azole
- Heteroaromatic compound
- Imidazole
- Tetrahydrofuran
- Secondary alcohol
- Secondary carboxylic acid amide
- Amino acid or derivatives
- Carboxamide group
- Organoheterocyclic compound
- Carboxylic acid derivative
- Alkylthiol
- Oxacycle
- Azacycle
- Organic oxide
- Organooxygen compound
- Organosulfur compound
- Carbonyl group
- Organopnictogen compound
- Organic nitrogen compound
- Organic oxygen compound
- Hydrocarbon derivative
- Alcohol
- Organonitrogen compound
- Primary amine
- Amine
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Charge | 0 |
|---|
| Melting point | Not Available |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Water Solubility | Not Available | PhysProp | | LogP | Not Available | PhysProp |
|
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations | Not Available |
|---|
| Organoleptic Properties | Not Available |
|---|
| SMPDB Pathways | |
|---|
| KEGG Pathways | | Pantothenate and CoA biosynthesis | ec00770 | 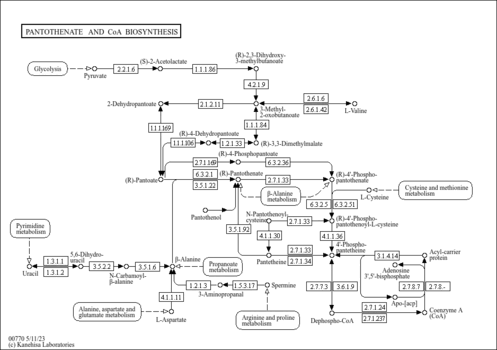 | | beta-Alanine metabolism | ec00410 |  |
|
|---|
| SMPDB Reactions | Not Available |
|---|
| KEGG Reactions | Not Available |
|---|
| Concentrations |
|---|
| Intracellular Concentrations | Not Available |
|---|
| Extracellular Concentrations | Not Available |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_5) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_6) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_7) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_8) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_9) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_5) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_6) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_7) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_8) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_9) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_10) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_11) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_12) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_13) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_14) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_15) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_16) - 70eV, Positive | Not Available | JSpectraViewer | | LC-MS/MS | LC-MS/MS Spectrum - 30V, Negative | splash10-052r-3019602000-8f7d5837950327bad269 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - 30V, Positive | splash10-000i-0010009000-a1396569e10326789a1c | JSpectraViewer | MoNA | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0040-2910618000-58e100b1447a771ecd7c | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-001i-1910200000-ba57f132dee9798389fa | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a7i-3900000000-d382c7ac6874be7ea151 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000i-0000009000-6bb1727ae8d28e18aad4 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-001i-2900023000-137fd3830c40fcb18b8f | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-05r0-4823906000-d707e02b69f32a1d64dd | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-2930102000-19d2796dee5d14788e39 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-3930000000-d4541feacee9a6c7a4e3 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-4900000000-1307cbce80b215d981b9 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-0800009000-9da226b219deabf5a54d | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-01p9-3960003000-62455c2483de897db0c6 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03di-1190000000-82c68976d04b3fab53ee | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer |
|
|---|
| References |
|---|
| References: | - Herrgard, M. J., Swainston, N., Dobson, P., Dunn, W. B., Arga, K. Y., Arvas, M., Bluthgen, N., Borger, S., Costenoble, R., Heinemann, M., Hucka, M., Le Novere, N., Li, P., Liebermeister, W., Mo, M. L., Oliveira, A. P., Petranovic, D., Pettifer, S., Simeonidis, E., Smallbone, K., Spasic, I., Weichart, D., Brent, R., Broomhead, D. S., Westerhoff, H. V., Kirdar, B., Penttila, M., Klipp, E., Palsson, B. O., Sauer, U., Oliver, S. G., Mendes, P., Nielsen, J., Kell, D. B. (2008). "A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology." Nat Biotechnol 26:1155-1160.18846089
- Cartwright, J. L., Gasmi, L., Spiller, D. G., McLennan, A. G. (2000). "The Saccharomyces cerevisiae PCD1 gene encodes a peroxisomal nudix hydrolase active toward coenzyme A and its derivatives." J Biol Chem 275:32925-32930.10922370
- Hampsey, D. M., Kohlhaw, G. B. (1981). "Inactivation of yeast alpha-isopropylmalate synthase by CoA. Antagonism between CoA and adenylates and the mechanism of CoA inactivation." J Biol Chem 256:3791-3796.7012143
|
|---|
| Synthesis Reference: | Not Available |
|---|
| External Links: | |
|---|


