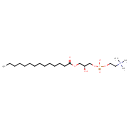| Identification |
|---|
| YMDB ID | YMDB01175 |
|---|
| Name | PC(14:0/0:0) |
|---|
| Species | Saccharomyces cerevisiae |
|---|
| Strain | Brewer's yeast |
|---|
| Description | LysoPC(14:0) is a lysophospholipid (LyP). It is a monoglycerophospholipid in which a phosphorylcholine moiety occupies a glycerol substitution site. Lysophosphatidylcholines can have different combinations of fatty acids of varying lengths and saturation attached at the C-1 (sn-1) position. Fatty acids containing 16, 18 and 20 carbons are the most common. LysoPC(14:0), in particular, consists of one chain of myristic acid at the C-1 position. Lysophosphatidylcholine is found in small amounts in most tissues. It is formed by hydrolysis of phosphatidylcholine by the enzyme phospholipase A2, as part of the de-acylation/re-acylation cycle that controls its overall molecular species composition. It can also be formed inadvertently during extraction of lipids from tissues if the phospholipase is activated by careless handling. |
|---|
| Structure | |
|---|
| Synonyms | - 1-acyl-2-lyso-phosphatidylcholine
- 1-Acyl-sn-glycero-3-phosphocholine
- 1-acyl-sn-glycero-3-phosphocholines
- 1-Acyl-sn-glycerol-3-phosphocholine
- 1-Acylglycerophosphocholine
- 1-myristoyl-glycero-3-phosphocholine
- 2-Lysolecithin
- 2-Lysophosphatidylcholine
- alpha-Acylglycerophosphocholine
- LPC
- LPC(14:0)
- LPC(14:0/0:0)
- LyPC(14:0)
- LyPC(14:0/0:0)
- LysoPC(14:0)
- LysoPC(14:0/0:0)
- Lysophosphatidylcholine(14:0)
- Lysophosphatidylcholine(14:0/0:0)
- 1-C14:0-Lysophosphatidylcholine betaine
- 1-Myristoyl-sn-glycero-3-phosphocholine betaine
- 1-Tetradecanoyl-sn-glycero-3-phosphocholine
- 1-Tetradecanoyl-sn-glycero-3-phosphocholine betaine
- PC(14:0/0:0)
- 1-tetradecanoyl-glycero-3-phosphocholine
- 1-Myristoylglycerophosphocholine
- 1-Tetradecanoylglycerophosphocholine
- 1-Myristoyl-GPC
- 1-Myristoyl-lysophosphatidylcholine
- 1-Myristoyl-sn-glycero-3-phosphocholine
- GPC(14:0)
- GPC(14:0/0:0)
|
|---|
| CAS number | Not Available |
|---|
| Weight | Average: 467.5769
Monoisotopic: 467.301189343 |
|---|
| InChI Key | VXUOFDJKYGDUJI-OAQYLSRUSA-N |
|---|
| InChI | InChI=1S/C22H46NO7P/c1-5-6-7-8-9-10-11-12-13-14-15-16-22(25)28-19-21(24)20-30-31(26,27)29-18-17-23(2,3)4/h21,24H,5-20H2,1-4H3/t21-/m1/s1 |
|---|
| IUPAC Name | (2-{[(2R)-2-hydroxy-3-(tetradecanoyloxy)propyl phosphono]oxy}ethyl)trimethylazanium |
|---|
| Traditional IUPAC Name | (2-{[(2R)-2-hydroxy-3-(tetradecanoyloxy)propyl phosphono]oxy}ethyl)trimethylazanium |
|---|
| Chemical Formula | C22H46NO7P |
|---|
| SMILES | [H][C@@](O)(COC(=O)CCCCCCCCCCCCC)COP([O-])(=O)OCC[N+](C)(C)C |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as 1-acyl-sn-glycero-3-phosphocholines. These are glycerophosphocholines in which the glycerol is esterified with a fatty acid at O-1 position, and linked at position 3 to a phosphocholine. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Glycerophospholipids |
|---|
| Sub Class | Glycerophosphocholines |
|---|
| Direct Parent | 1-acyl-sn-glycero-3-phosphocholines |
|---|
| Alternative Parents | |
|---|
| Substituents | - 1-acyl-sn-glycero-3-phosphocholine
- Phosphocholine
- Fatty acid ester
- Dialkyl phosphate
- Organic phosphoric acid derivative
- Phosphoric acid ester
- Alkyl phosphate
- Fatty acyl
- Tetraalkylammonium salt
- Quaternary ammonium salt
- Secondary alcohol
- Carboxylic acid ester
- Carboxylic acid derivative
- Monocarboxylic acid or derivatives
- Organic oxide
- Organooxygen compound
- Organonitrogen compound
- Organic nitrogen compound
- Alcohol
- Organic oxygen compound
- Organopnictogen compound
- Carbonyl group
- Organic salt
- Amine
- Hydrocarbon derivative
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Charge | 0 |
|---|
| Melting point | Not Available |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Water Solubility | Not Available | PhysProp | | LogP | Not Available | PhysProp |
|
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations | Not Available |
|---|
| Organoleptic Properties | Not Available |
|---|
| SMPDB Pathways | | Glycerophospholipid metabolism | PW002493 | 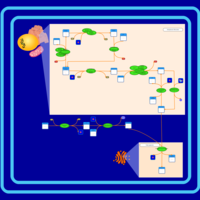 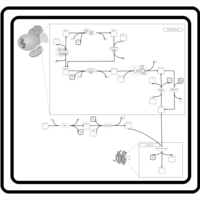 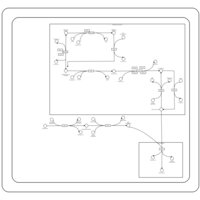 | | Lysolipid incorporation into ER | PW002532 | 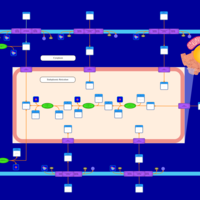 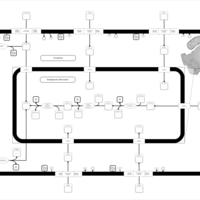 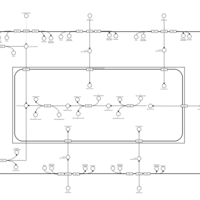 | | Lysolipid incorporation into ER PC(14:0/14:0) | PW002783 | 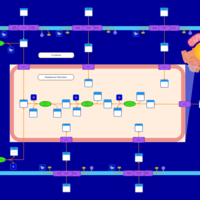 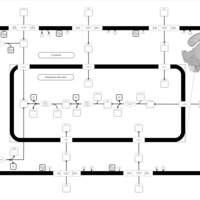 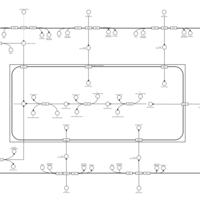 | | Lysolipid incorporation into ER PC(16:1(11Z)/16:1(11Z)) | PW002786 | 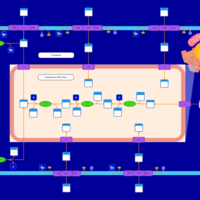 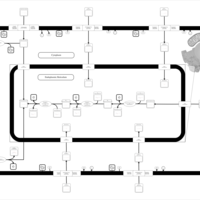 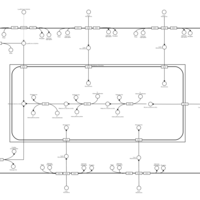 | | Lysolipid incorporation into ER PC(18:2(9Z,11Z)/18:2(9Z,11Z)) | PW002789 | 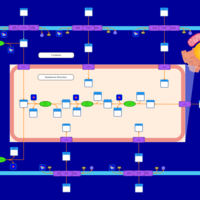 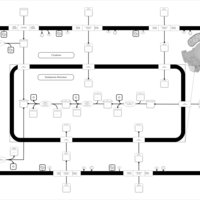 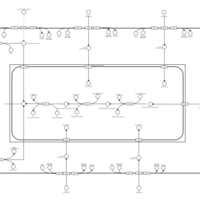 |
|
|---|
| KEGG Pathways | | Glycerophospholipid metabolism | ec00564 |  |
|
|---|
| SMPDB Reactions |
|
|---|
| KEGG Reactions | Not Available |
|---|
| Concentrations |
|---|
| Intracellular Concentrations | Not Available |
|---|
| Extracellular Concentrations | Not Available |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-0230-9411000000-7a6b8e8fd1e0609b417e | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0006-0000900000-c557e23aa67a6a68e471 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-001l-0000900000-89cc0e21e01554e310d6 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0pcr-1617900000-231c7760e10f0c81e04b | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udi-0000390000-b1814b140293d64c3417 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0fb9-0090160000-48fcbf8694eed693a211 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-0090110000-c40c1368f5389cd115f2 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0gb9-0000900000-534f405ea851fa87d3e5 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00lr-0900600000-9a2a86225de12cddb6bd | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0ue9-0910400000-63d03888aa82a44da515 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014i-0030900000-648928c7aa526a1c5514 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-0090100000-6eecefd72f10fda6c57a | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-0090000000-aaa1334bcdf77ff34dd1 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00di-0000900000-56c0d0685c46c9ba6fe3 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-01i0-0010900000-50babd7089582cb56ec8 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0frl-0390400000-26a01ba41c58e985ad7c | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer |
|
|---|
| References |
|---|
| References: | - Ejsing, C. S., Sampaio, J. L., Surendranath, V., Duchoslav, E., Ekroos, K., Klemm, R. W., Simons, K., Shevchenko, A. (2009). "Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry." Proc Natl Acad Sci U S A 106:2136-2141.19174513
|
|---|
| Synthesis Reference: | Not Available |
|---|
| External Links: | |
|---|