| Identification |
|---|
| YMDB ID | YMDB00884 |
|---|
| Name | Linoleic acid |
|---|
| Species | Saccharomyces cerevisiae |
|---|
| Strain | Baker's yeast |
|---|
| Description | Linoelaidic acid, also known as linoelaidate, belongs to the class of organic compounds known as lineolic acids and derivatives. These are derivatives of lineolic acid. Lineolic acid is a polyunsaturated omega-6 18 carbon long fatty acid, with two CC double bonds at the 9- and 12-positions. Based on a literature review a significant number of articles have been published on Linoelaidic acid. |
|---|
| Structure | |
|---|
| Synonyms | - (9Z,12Z)-9,12-Octadecadienoate
- (9Z,12Z)-9,12-Octadecadienoic acid
- (Z,Z)-9,12-Octadecadienoate
- (Z,Z)-9,12-Octadecadienoic acid
- 9-cis,12-cis-Linoleate
- 9-cis,12-cis-Linoleic acid
- 9Z,12Z-Linoleate
- 9Z,12Z-Linoleic acid
- 9Z,12Z-Octadecadienoate
- 9Z,12Z-Octadecadienoic acid
- all-cis-9,12-Octadecadienoate
- all-cis-9,12-Octadecadienoic acid
- cis-9,cis-12-Octadecadienoate
- cis-9,cis-12-Octadecadienoic acid
- cis-D9,12-Octadecadienoate
- cis-D9,12-Octadecadienoic acid
- cis,cis-Linoleate
- cis,cis-Linoleic acid
- Linolate
- Linoleate
- Linoleic acid
- Linolic acid
- 18:2, N-6,9 all-trans
- 9E,12E-Octadecadienoic acid
- C18:2, N-6,9 all-trans
- trans-9,trans-12-Linoleic acid
- 9E,12E-Octadecadienoate
- trans-9,trans-12-Linoleate
- Linoelaidate
- (9E,12E)-9,12-Octadecadienoate
- (9E,12E)-9,12-Octadecadienoic acid
- 9,12-Octadecadienoic acid
- Linoleic acid, (Z,Z)-isomer, 14C-labeled
- Linolelaidic acid
- cis,cis-9,12-Octadecadienoic acid
- Linoleic acid, (e,e)-isomer
- Linoleic acid, (Z,Z)-isomer
- Linoleic acid, ammonium salt, (Z,Z)-isomer
- Linoleic acid, potassium salt, (Z,Z)-isomer
- Linoelaidic acid, (e,Z)-isomer
- 9 trans,12 trans Octadecadienoic acid
- Linoleic acid, sodium salt, (e,e)-isomer
- Linoleic acid, sodium salt, (Z,Z)-isomer
- Acid, 9,12-octadecadienoic
- trans,trans-9,12-Octadecadienoic acid
- Linoleic acid, (Z,e)-isomer
- 9,12 Octadecadienoic acid
- Linoleic acid, calcium salt, (Z,Z)-isomer
- 9-trans,12-trans-Octadecadienoic acid
|
|---|
| CAS number | 60-33-3 |
|---|
| Weight | Average: 280.4455
Monoisotopic: 280.240230268 |
|---|
| InChI Key | OYHQOLUKZRVURQ-AVQMFFATSA-N |
|---|
| InChI | InChI=1S/C18H32O2/c1-2-3-4-5-6-7-8-9-10-11-12-13-14-15-16-17-18(19)20/h6-7,9-10H,2-5,8,11-17H2,1H3,(H,19,20)/b7-6+,10-9+ |
|---|
| IUPAC Name | (9E,12E)-octadeca-9,12-dienoic acid |
|---|
| Traditional IUPAC Name | linoelaidic acid |
|---|
| Chemical Formula | C18H32O2 |
|---|
| SMILES | [H]OC(=O)C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])=C([H])C([H])([H])C([H])=C([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])[H] |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as lineolic acids and derivatives. These are derivatives of lineolic acid. Lineolic acid is a polyunsaturated omega-6 18 carbon long fatty acid, with two CC double bonds at the 9- and 12-positions. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Fatty Acyls |
|---|
| Sub Class | Lineolic acids and derivatives |
|---|
| Direct Parent | Lineolic acids and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Octadecanoid
- Long-chain fatty acid
- Fatty acid
- Unsaturated fatty acid
- Straight chain fatty acid
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Carboxylic acid derivative
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Carbonyl group
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State | Liquid |
|---|
| Charge | 0 |
|---|
| Melting point | -8.5 °C |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Water Solubility | Not Available | PhysProp | | LogP | 7.05 [SANGSTER (1993)] | PhysProp |
|
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations | - cell envelope
- mitochondrion
- extracellular
- endoplasmic reticulum
- lipid particle
- cytoplasm
|
|---|
| Organoleptic Properties | |
|---|
| SMPDB Pathways | | Biosynthesis of unsaturated fatty acids | PW002403 |    |
|
|---|
| KEGG Pathways | | Biosynthesis of unsaturated fatty acids | ec01040 |  | | Linoleic acid metabolism | ec00591 | 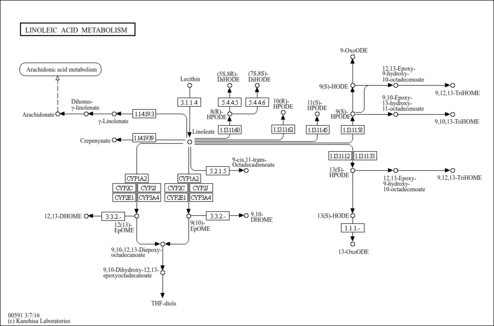 |
|
|---|
| SMPDB Reactions | Not Available |
|---|
| KEGG Reactions | |
|---|
| Concentrations |
|---|
| Intracellular Concentrations | Not Available |
|---|
| Extracellular Concentrations | Not Available |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0007-9750000000-50d69948d56dd2ba6e42 | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-0079-9631000000-cc93c24bbf14d81ae9b6 | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | JSpectraViewer | | LC-MS/MS | LC-MS/MS Spectrum - ESI-TOF 10V, Negative | splash10-0udi-0009000000-4dd23c77f8e48c0ab9ec | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - ESI-TOF 20V, Negative | splash10-0udi-0009000000-4dd23c77f8e48c0ab9ec | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - ESI-TOF 10V, Negative | splash10-004i-0092000000-b8aabbdc61f9b89ac359 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - ESI-TOF , Negative | splash10-004i-0092000000-b8aabbdc61f9b89ac359 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - ESI-TOF 30V, Negative | splash10-004i-0092000000-b8aabbdc61f9b89ac359 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - ESI-TOF 10V, Negative | splash10-004i-0090000000-dd6cf3859995c0ba8be8 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - ESI-TOF 20V, Negative | splash10-004i-0090000000-fdabfebc98e900a3a52b | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - ESI-TOF 10V, Negative | splash10-004i-0092000000-b8aabbdc61f9b89ac359 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - ESI-TOF , Negative | splash10-004i-0091000000-d8e32f26b102cfd39457 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - ESI-TOF 30V, Negative | splash10-004i-0090000000-e1aa5a6fc5d53c0d0cb6 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-TOF , negative | splash10-004i-0090000000-dd6cf3859995c0ba8be8 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-TOF , negative | splash10-004i-0090000000-fdabfebc98e900a3a52b | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-TOF , negative | splash10-004i-0090000000-e1aa5a6fc5d53c0d0cb6 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-004i-0090000000-fdabfebc98e900a3a52b | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - 10V, Positive | splash10-004i-0090000000-dd6cf3859995c0ba8be8 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - 30V, Positive | splash10-004i-0090000000-e1aa5a6fc5d53c0d0cb6 | JSpectraViewer | MoNA | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-01q9-0190000000-76fe39ad6bbee3f769e7 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0239-5690000000-9af1d7ecbc9ccf182d61 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-006x-8930000000-0032f23abecb3b362ff2 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-0090000000-b26f2ec0d32875acf897 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004r-1090000000-08e28514e79444592bb5 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4l-9230000000-d3f27ea65196f222f6cd | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-0090000000-03fd7d53d39127d02766 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-01t9-1090000000-e5b6404e73269d5a2528 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9210000000-da81d6d1d7b1e626e905 | JSpectraViewer |
|
|---|
| References |
|---|
| References: | - Herrgard, M. J., Swainston, N., Dobson, P., Dunn, W. B., Arga, K. Y., Arvas, M., Bluthgen, N., Borger, S., Costenoble, R., Heinemann, M., Hucka, M., Le Novere, N., Li, P., Liebermeister, W., Mo, M. L., Oliveira, A. P., Petranovic, D., Pettifer, S., Simeonidis, E., Smallbone, K., Spasic, I., Weichart, D., Brent, R., Broomhead, D. S., Westerhoff, H. V., Kirdar, B., Penttila, M., Klipp, E., Palsson, B. O., Sauer, U., Oliver, S. G., Mendes, P., Nielsen, J., Kell, D. B. (2008). "A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology." Nat Biotechnol 26:1155-1160.18846089
|
|---|
| Synthesis Reference: | Walborsky, Harry M.; Davis, Robert H.; Howton, David R. A total synthesis of linoleic acid. Journal of the American Chemical Society (1951), 73 2590-4. |
|---|
| External Links: | |
|---|


