| Identification |
|---|
| YMDB ID | YMDB00770 |
|---|
| Name | taurocholic acid |
|---|
| Species | Saccharomyces cerevisiae |
|---|
| Strain | Baker's yeast |
|---|
| Description | Taurocholic acid, also known as taurocholate or choloyl-taurine, belongs to the class of organic compounds known as trihydroxy bile acids, alcohols and derivatives. These are prenol lipids structurally characterized by a bile acid or alcohol which bears three hydroxyl groups. Taurocholic acid is a very hydrophobic molecule, practically insoluble (in water), and relatively neutral. |
|---|
| Structure | |
|---|
| Synonyms | - 3a,7a,12a-Trihydroxy-5b-cholanic acid-24-taurine
- 3alpha,7alpha,12alpha-trihydroxy-5beta-cholanic acid 24-taurine
- bile acid
- Cholaic acid
- Cholic acid taurine conjugate
- Choloyl-taurine
- cholyltaurine
- n-choloyl-Taurine
- N-Choloyltaurine
- Taurine, N-choloyl-
- taurocholate
- Taurocholic acid
- 3a,7a,12a-Trihydroxy-5b-cholanate 24-taurine
- 3a,7a,12a-Trihydroxy-5b-cholanic acid 24-taurine
- 3alpha,7alpha,12alpha-Trihydroxy-5beta-cholanate 24-taurine
- 3Α,7α,12α-trihydroxy-5β-cholanate 24-taurine
- 3Α,7α,12α-trihydroxy-5β-cholanic acid 24-taurine
- Cholate taurine conjugate
- Cholic acid taurine conjugic acid
- Cholaate
- Taurine cholate
- Taurocholic acid, (7 beta)-isomer
- Taurocholic acid, (5 alpha)-isomer
|
|---|
| CAS number | 81-24-3 |
|---|
| Weight | Average: 515.703
Monoisotopic: 515.291673489 |
|---|
| InChI Key | WBWWGRHZICKQGZ-HZAMXZRMSA-N |
|---|
| InChI | InChI=1S/C26H45NO7S/c1-15(4-7-23(31)27-10-11-35(32,33)34)18-5-6-19-24-20(14-22(30)26(18,19)3)25(2)9-8-17(28)12-16(25)13-21(24)29/h15-22,24,28-30H,4-14H2,1-3H3,(H,27,31)(H,32,33,34)/t15-,16+,17-,18-,19+,20+,21-,22+,24+,25+,26-/m1/s1 |
|---|
| IUPAC Name | 2-[(4R)-4-[(1S,2S,5R,7S,9R,10R,11S,14R,15R,16S)-5,9,16-trihydroxy-2,15-dimethyltetracyclo[8.7.0.0^{2,7}.0^{11,15}]heptadecan-14-yl]pentanamido]ethane-1-sulfonic acid |
|---|
| Traditional IUPAC Name | 2-[(4R)-4-[(1S,2S,5R,7S,9R,10R,11S,14R,15R,16S)-5,9,16-trihydroxy-2,15-dimethyltetracyclo[8.7.0.0^{2,7}.0^{11,15}]heptadecan-14-yl]pentanamido]ethanesulfonic acid |
|---|
| Chemical Formula | C26H45NO7S |
|---|
| SMILES | [H][C@@]1(CC[C@@]2([H])[C@]3([H])[C@H](O)C[C@]4([H])C[C@H](O)CC[C@]4(C)[C@@]3([H])C[C@H](O)[C@]12C)[C@H](C)CCC(=O)NCCS(O)(=O)=O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as trihydroxy bile acids, alcohols and derivatives. These are prenol lipids structurally characterized by a bile acid or alcohol which bears three hydroxyl groups. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Steroids and steroid derivatives |
|---|
| Sub Class | Bile acids, alcohols and derivatives |
|---|
| Direct Parent | Trihydroxy bile acids, alcohols and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Trihydroxy bile acid, alcohol, or derivatives
- 3-hydroxysteroid
- 12-hydroxysteroid
- Hydroxysteroid
- 3-alpha-hydroxysteroid
- 7-hydroxysteroid
- Cyclic alcohol
- Organic sulfonic acid or derivatives
- Organosulfonic acid or derivatives
- Organosulfonic acid
- Sulfonyl
- Alkanesulfonic acid
- Secondary alcohol
- Carboximidic acid
- Polyol
- Carboximidic acid derivative
- Organic 1,3-dipolar compound
- Propargyl-type 1,3-dipolar organic compound
- Alcohol
- Organosulfur compound
- Organooxygen compound
- Organonitrogen compound
- Organopnictogen compound
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organic nitrogen compound
- Aliphatic homopolycyclic compound
|
|---|
| Molecular Framework | Aliphatic homopolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Charge | 0 |
|---|
| Melting point | 125 °C |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Water Solubility | Not Available | PhysProp | | LogP | Not Available | PhysProp |
|
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations | |
|---|
| Organoleptic Properties | Not Available |
|---|
| SMPDB Pathways | Not Available |
|---|
| KEGG Pathways | | Taurine and hypotaurine metabolism | ec00430 | 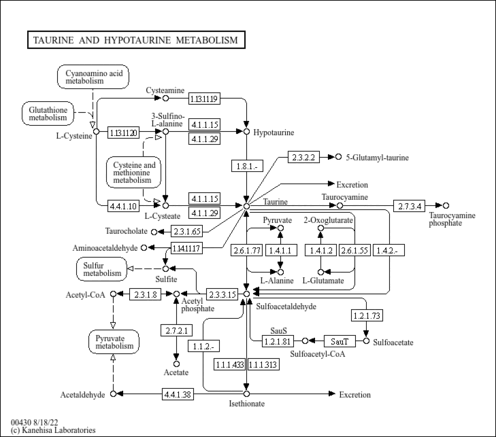 |
|
|---|
| SMPDB Reactions | Not Available |
|---|
| KEGG Reactions | |
|---|
| Concentrations |
|---|
| Intracellular Concentrations | Not Available |
|---|
| Extracellular Concentrations | Not Available |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_5) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_5) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_6) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_7) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_8) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_9) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_10) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_1) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_2) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_3) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_4) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_5) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_6) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_7) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_8) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_9) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_10) - 70eV, Positive | Not Available | JSpectraViewer | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-03di-0000090000-911b2f8adf5d8a253a26 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-03di-0000090000-229891181661857b574b | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-03di-0000090000-e3b432e2a36b91fb1106 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-03di-0000090000-afa59839c0e5c1b1f875 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-03di-0000090000-b29692cfc9f66c4205c5 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-IT , negative | splash10-0gvk-0019610000-6cd348047dc06e3abb7e | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-014i-0010390000-b424218e255bf3091fda | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-0a4i-0191200000-4fddfc0190213d6555ee | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-08i9-0590000000-c210cb519c7e4192622e | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-0w29-0900000000-42467048740170d1f693 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-IT , positive | splash10-0002-0000940000-6c3a6cb6cef0d74f2139 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-03di-0000090000-21f22221a28d374a2518 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - 10V, Positive | splash10-03di-0000900000-bc321572fda51c26ea4b | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - 40V, Positive | splash10-004i-2961000000-2ad87ae8f70b4deb4613 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - 35V, Negative | splash10-03di-0030090000-4366a02ee69b6b2b9a47 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-03di-0114900000-263e4adb5d4c8f0a9166 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-03di-0000900000-ffd27384b632f9e9db56 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - 40V, Positive | splash10-01p9-0389300000-86e8186c77b1ec43a6b7 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - 10V, Positive | splash10-014j-0000960000-cd066ae54dc0cc5a21fe | JSpectraViewer | MoNA | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00ea-0601910000-36005a0ab81d9a5c22c7 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00di-2901200000-0277ee57e5a77bb559ed | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-006x-8903200000-6e18275cb262330c1d11 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03dj-3102890000-14b58b657187799f4abd | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-01qa-6404930000-b60c0ef5b779fc055945 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-000x-9101100000-30f5c6ad01e5a5d282da | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 2D NMR | [1H,13C] 2D NMR Spectrum | Not Available | JSpectraViewer |
|
|---|
| References |
|---|
| References: | - Herrgard, M. J., Swainston, N., Dobson, P., Dunn, W. B., Arga, K. Y., Arvas, M., Bluthgen, N., Borger, S., Costenoble, R., Heinemann, M., Hucka, M., Le Novere, N., Li, P., Liebermeister, W., Mo, M. L., Oliveira, A. P., Petranovic, D., Pettifer, S., Simeonidis, E., Smallbone, K., Spasic, I., Weichart, D., Brent, R., Broomhead, D. S., Westerhoff, H. V., Kirdar, B., Penttila, M., Klipp, E., Palsson, B. O., Sauer, U., Oliver, S. G., Mendes, P., Nielsen, J., Kell, D. B. (2008). "A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology." Nat Biotechnol 26:1155-1160.18846089
- Rebbeor, J. F., Connolly, G. C., Ballatori, N. (2002). "Inhibition of Mrp2- and Ycf1p-mediated transport by reducing agents: evidence for GSH transport on rat Mrp2." Biochim Biophys Acta 1559:171-178.11853683
|
|---|
| Synthesis Reference: | Schersten, Tore; Bjorntorp, Per; Ekdahi, Per H.; Bjorkerud, Soren. Synthesis of taurocholic and glycocholic acids by preparations of human liver. II. An analysis of the stimulating effect of the L fraction. Biochimica et Biophysica Acta, General Sub |
|---|
| External Links: | |
|---|

