| Identification |
|---|
| YMDB ID | YMDB00674 |
|---|
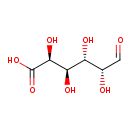
| Name | aldehydo-D-galacturonate |
|---|
| Species | Saccharomyces cerevisiae |
|---|
| Strain | Baker's yeast |
|---|
| Description | Galacturonic acid, also known as D-galacturonate or sodium pectate, belongs to the class of organic compounds known as glucuronic acid derivatives. Glucuronic acid derivatives are compounds containing a glucuronic acid moiety (or a derivative), which consists of a glucose moiety with the C6 carbon oxidized to a carboxylic acid. Galacturonic acid exists in all living species, ranging from bacteria to plants to humans. Based on a literature review a significant number of articles have been published on Galacturonic acid. |
|---|
| Structure | |
|---|
| Synonyms | - D-Galactopyranuronic acid
- D-Galacturonate
- D-Galacturonic acid
- Galactopyranuronic acid, D-
- Galacturonic acid, D-
- hexuronic acid
- (2S,3R,4S,5R)-2,3,4,5-Tetrahydroxy-6-oxohexanoic acid
- (2S,3R,4S,5R)-2,3,4,5-Tetrahydroxy-6-oxohexanoate
- Galacturonate
- DL-Galacturonic acid
- Galacturonic acid, (D)-isomer
- Galacturonic acid, (alpha-D)-isomer
- Galacturonic acid, calcium, sodium salt, (D)-isomer
- Galacturonic acid, monosodium salt, (D)-isomer
- Aldehydo-D-galacturonate
- Polygalacturonic acid, aluminum salt
- Sodium pectate
- Galacturonan
- Homogalacturonan
- Pectic acid
- Polygalacturonic acid homopolymer
- Polygalacturonic acid, sulfated
- Calcium polygalacturonate
- Pectate
- Polygalacturonic acid
- Polygalacturonic acid, calcium salt
- Polygalacturonic acid, homopolymer sodium salt
- Sodium polygalacturonate
- Anhydrogalacturonic acid
- Calcium pectate
- Polygalacturonic acid, homopolymer (D)-isomer
- Aldehydo-D-galacturonic acid
- (DL)-Galacturonic acid
- (D)-Galacturonic acid
- Galacturonic acid
|
|---|
| CAS number | 685-73-4 |
|---|
| Weight | Average: 194.1394
Monoisotopic: 194.042652674 |
|---|
| InChI Key | IAJILQKETJEXLJ-RSJOWCBRSA-N |
|---|
| InChI | InChI=1S/C6H10O7/c7-1-2(8)3(9)4(10)5(11)6(12)13/h1-5,8-11H,(H,12,13)/t2-,3+,4+,5-/m0/s1 |
|---|
| IUPAC Name | (2S,3R,4S,5R)-2,3,4,5-tetrahydroxy-6-oxohexanoic acid |
|---|
| Traditional IUPAC Name | aldehydo-D-galacturonic acid |
|---|
| Chemical Formula | C6H10O7 |
|---|
| SMILES | [H]C(=O)[C@H](O)[C@@H](O)[C@@H](O)[C@H](O)C(O)=O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as glucuronic acid derivatives. Glucuronic acid derivatives are compounds containing a glucuronic acid moiety (or a derivative), which consists of a glucose moiety with the C6 carbon oxidized to a carboxylic acid. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | Glucuronic acid derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Glucuronic acid or derivatives
- Hexose monosaccharide
- Medium-chain hydroxy acid
- Medium-chain fatty acid
- Beta-hydroxy acid
- Hydroxy fatty acid
- Monosaccharide
- Fatty acyl
- Hydroxy acid
- Fatty acid
- Alpha-hydroxy acid
- Beta-hydroxy aldehyde
- Alpha-hydroxyaldehyde
- Secondary alcohol
- Polyol
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Carboxylic acid derivative
- Aldehyde
- Carbonyl group
- Alcohol
- Organic oxide
- Hydrocarbon derivative
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Charge | 0 |
|---|
| Melting point | 166 °C |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Water Solubility | Not Available | PhysProp | | LogP | Not Available | PhysProp |
|
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations | |
|---|
| Organoleptic Properties | Not Available |
|---|
| SMPDB Pathways | | Amino sugar and nucleotide sugar metabolism | PW002413 | 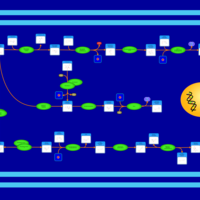 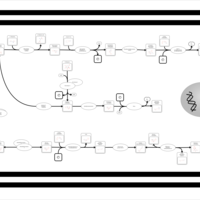 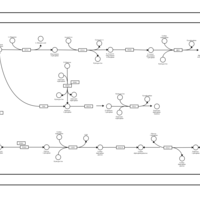 | | Starch and sucrose metabolism | PW002481 | 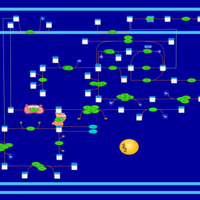 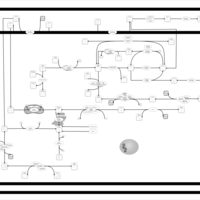 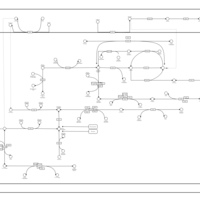 |
|
|---|
| KEGG Pathways | | Amino sugar and nucleotide sugar metabolism | ec00520 |  | | Pentose and glucuronate interconversions | ec00040 | 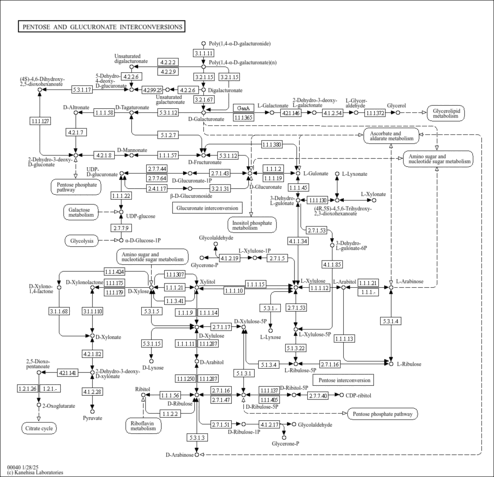 | | Starch and sucrose metabolism | ec00500 |  |
|
|---|
| SMPDB Reactions | Not Available |
|---|
| KEGG Reactions | |
|---|
| Concentrations |
|---|
| Intracellular Concentrations | Not Available |
|---|
| Extracellular Concentrations | Not Available |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-000i-9800000000-1d25ab641e968717daf1 | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (5 TMS) - 70eV, Positive | splash10-00n0-3139450000-1762a4c61557c689a607 | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | JSpectraViewer | | LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-03e9-0900000000-04b6c8cb4b4b2c077202 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-0lk9-9700000000-4260c2e124c927041b62 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-004i-9000000000-650b5bb9db964b1c3e87 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - , negative | splash10-000i-9410000000-180d0e747dbcb5b21b01 | JSpectraViewer | MoNA | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-056s-2900000000-86606b8382ffe403bc8e | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a4i-9500000000-af67bc97552641417bee | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4i-9100000000-205a7cff1e298283a080 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-05p9-9700000000-ee070fab487c9f3e49b7 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4r-9400000000-284825e422ba9585f17b | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-9100000000-64d4f9d758e2a4eeba68 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-01y5-7900000000-df218a4e340a0ff668c1 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03kl-9100000000-d22d4d2644d77203331d | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-08fu-9000000000-93301767b758d212114f | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00fr-9300000000-77aa1619935fe785e397 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a6r-9000000000-da4be7e32a829503c021 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a6r-9000000000-c1c9308cd925f43d015f | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 2D NMR | [1H,13C] 2D NMR Spectrum | Not Available | JSpectraViewer |
|
|---|
| References |
|---|
| References: | - Herrgard, M. J., Swainston, N., Dobson, P., Dunn, W. B., Arga, K. Y., Arvas, M., Bluthgen, N., Borger, S., Costenoble, R., Heinemann, M., Hucka, M., Le Novere, N., Li, P., Liebermeister, W., Mo, M. L., Oliveira, A. P., Petranovic, D., Pettifer, S., Simeonidis, E., Smallbone, K., Spasic, I., Weichart, D., Brent, R., Broomhead, D. S., Westerhoff, H. V., Kirdar, B., Penttila, M., Klipp, E., Palsson, B. O., Sauer, U., Oliver, S. G., Mendes, P., Nielsen, J., Kell, D. B. (2008). "A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology." Nat Biotechnol 26:1155-1160.18846089
|
|---|
| Synthesis Reference: | Not Available |
|---|
| External Links: | |
|---|