| Identification |
|---|
| YMDB ID | YMDB00287 |
|---|
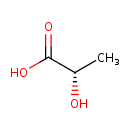
| Name | D-Lactic acid |
|---|
| Species | Saccharomyces cerevisiae |
|---|
| Strain | Baker's yeast |
|---|
| Description | L-Lactic acid, also known as L-lactate or L-milchsaeure, belongs to the class of organic compounds known as alpha hydroxy acids and derivatives. These are organic compounds containing a carboxylic acid substituted with a hydroxyl group on the adjacent carbon. L-Lactic acid is an extremely weak basic (essentially neutral) compound (based on its pKa). L-Lactic acid is a potentially toxic compound. |
|---|
| Structure | |
|---|
| Synonyms | - (-)-Lactate
- (-)-Lactic acid
- (D)-(-)-Lactic acid
- (R)-(-)-Lactate
- (R)-(-)-Lactic acid
- (R)-2-Hydroxypropanoate
- (R)-2-Hydroxypropanoic acid
- (R)-2-Hydroxypropionate
- (R)-2-Hydroxypropionic acid
- (R)-2-Hydroxypropionsaeure
- (R)-a-Hydroxypropionate
- (R)-a-Hydroxypropionic acid
- (R)-alpha-Hydroxypropionate
- (R)-alpha-Hydroxypropionic acid
- (R)-Lactate
- (R)-Lactic acid
- (R)-Milchsaeure
- 1-Lactic acid
- D-(-)-Lactate
- D-(-)-Lactic acid
- D-2-Hydroxypropanoate
- D-2-Hydroxypropanoic acid
- D-2-Hydroxypropionate
- D-2-Hydroxypropionic acid
- D-Lactate
- D-Lactic acid
- D-Milchsaeure
- delta-(-)-Lactate
- delta-(-)-Lactic acid
- delta-2-Hydroxypropanoate
- delta-2-Hydroxypropanoic acid
- delta-2-Hydroxypropionate
- delta-2-Hydroxypropionic acid
- delta-Lactate
- delta-Lactic acid
- DLA
- L-(+)-Lactate
- L-Lactic acid
- Lactic acid (D)
- Propanoic acid, 2-hydroxy-, (2R)-
- Propanoic acid, 2-hydroxy-, (R)-
- Propel
- Tisulac
- (+)-Lactic acid
- (S)-(+)-Lactic acid
- (S)-2-Hydroxypropanoic acid
- (S)-2-Hydroxypropionic acid
- L-(+)-alpha-Hydroxypropionic acid
- L-(+)-Lactic acid
- L-Milchsaeure
- L-Lactate
- (+)-Lactate
- (S)-(+)-Lactate
- (S)-2-Hydroxypropanoate
- (S)-2-Hydroxypropionate
- L-(+)-a-Hydroxypropionate
- L-(+)-a-Hydroxypropionic acid
- L-(+)-alpha-Hydroxypropionate
- L-(+)-Α-hydroxypropionate
- L-(+)-Α-hydroxypropionic acid
- (alpha)-Lactate
- (alpha)-Lactic acid
- (S)-(+)-2-Hydroxypropanoate
- (S)-(+)-2-Hydroxypropanoic acid
- (S)-2-Hydroxy-propanoate
- (S)-2-Hydroxy-propanoic acid
- (S)-Lactate
- (S)-Lactic acid
- 1-Hydroxyethane 1-carboxylate
- 1-Hydroxyethane 1-carboxylic acid
- 1-Hydroxyethanecarboxylate
- 1-Hydroxyethanecarboxylic acid
- 2-Hydroxypropanoate
- 2-Hydroxypropanoic acid
- 2-Hydroxypropionate
- a-Hydroxypropanoate
- a-Hydroxypropanoic acid
- a-Hydroxypropionate
- a-Hydroxypropionic acid
- alpha-Hydroxypropanoate
- alpha-Hydroxypropanoic acid
- alpha-Hydroxypropionate
- alpha-Hydroxypropionic acid
- L-(+)- Lactic acid
- L-2-Hydroxypropanoate
- L-2-Hydroxypropanoic acid
- Lactate
- Lactic acid
- Milk acid
- Sarcolactic acid
- 2-Hydroxypropionic acid
- D Lactic acid
- Lactate, ammonium
- 2 Hydroxypropanoic acid
- 2 Hydroxypropionic acid
- Ammonium lactate
- L Lactic acid
|
|---|
| CAS number | 10326-41-7 |
|---|
| Weight | Average: 90.0779
Monoisotopic: 90.031694058 |
|---|
| InChI Key | JVTAAEKCZFNVCJ-UWTATZPHSA-N |
|---|
| InChI | InChI=1S/C3H6O3/c1-2(4)3(5)6/h2,4H,1H3,(H,5,6)/t2-/m1/s1 |
|---|
| IUPAC Name | (2S)-2-hydroxypropanoic acid |
|---|
| Traditional IUPAC Name | (α)-lactate |
|---|
| Chemical Formula | C3H6O3 |
|---|
| SMILES | [H]OC(=O)[C@]([H])(O[H])C([H])([H])[H] |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as alpha hydroxy acids and derivatives. These are organic compounds containing a carboxylic acid substituted with a hydroxyl group on the adjacent carbon. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Hydroxy acids and derivatives |
|---|
| Sub Class | Alpha hydroxy acids and derivatives |
|---|
| Direct Parent | Alpha hydroxy acids and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Alpha-hydroxy acid
- Secondary alcohol
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Carboxylic acid derivative
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Carbonyl group
- Alcohol
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Charge | 0 |
|---|
| Melting point | 52.8 °C |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Water Solubility | Not Available | PhysProp | | LogP | Not Available | PhysProp |
|
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations | Not Available |
|---|
| Organoleptic Properties | |
|---|
| SMPDB Pathways | |
|---|
| KEGG Pathways | |
|---|
| SMPDB Reactions | |
|---|
| KEGG Reactions | |
|---|
| Concentrations |
|---|
| Intracellular Concentrations | Not Available |
|---|
| Extracellular Concentrations | Not Available |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (Non-derivatized) | splash10-00kb-0900000000-fb59ec16914501aa19ab | JSpectraViewer | MoNA | | GC-MS | GC-MS Spectrum - EI-B (Non-derivatized) | splash10-014j-0900000000-c4d9e12b4b0150eda54b | JSpectraViewer | MoNA | | GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-00kb-0900000000-fb59ec16914501aa19ab | JSpectraViewer | MoNA | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0006-9000000000-a3691f383d440fb00e1f | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-01b9-9620000000-f7faa7db9c1be3d9d975 | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_2) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_1) - 70eV, Positive | Not Available | JSpectraViewer | | LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Negative (Annotated) | splash10-000i-9000000000-1d5a5f55463acefb6fc7 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Negative (Annotated) | splash10-000m-9000000000-c07133799d8f43d4c75a | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Negative (Annotated) | splash10-000i-9000000000-ddb080250741e69ab137 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Negative | splash10-000i-9000000000-8f5d5eddb0b4b2a3b541 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Negative | splash10-000i-9000000000-3bcfc4cdc49230f15642 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Negative | splash10-0006-9000000000-48b511409f4f60cec04e | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Negative | splash10-0006-9000000000-365ea3c1bcfff2cab938 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 50V, Negative | splash10-0006-9000000000-9d0238aeeb837213e81d | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-000i-9000000000-8f5d5eddb0b4b2a3b541 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-000i-9000000000-3bcfc4cdc49230f15642 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-0006-9000000000-48b511409f4f60cec04e | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-0006-9000000000-365ea3c1bcfff2cab938 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-0006-9000000000-9d0238aeeb837213e81d | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - QTOF 30V, positive | splash10-014i-9000000000-046bb7f6cce9744388eb | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-000f-9000000000-1c6ea519686b2066052f | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - 35V, Negative | splash10-000i-9000000000-013763552dec660fac8c | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-0006-9000000000-bc4403a40640361e5a52 | JSpectraViewer | MoNA | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-006x-9000000000-5f417f4a6d08f0ab00ed | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00dm-9000000000-df7a94bb1a9cf6e78e1a | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-004j-9000000000-dc2a1b965287b9dfee9c | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000i-9000000000-c3686a681cc9bbf039e1 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-000i-9000000000-ddad20647c2ac56efd22 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00di-9000000000-b728b45617afcc6b67da | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000i-9000000000-b586cb8f053eb4465b4e | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-000i-9000000000-400a5f1c0dfcc32ef2bb | JSpectraViewer | | MS | Mass Spectrum (Electron Ionization) | splash10-002b-9000000000-50213d6b39ef9741c466 | JSpectraViewer | MoNA | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 2D NMR | [1H,1H] 2D NMR Spectrum | Not Available | JSpectraViewer | | 2D NMR | [1H,13C] 2D NMR Spectrum | Not Available | JSpectraViewer |
|
|---|
| References |
|---|
| References: | - UniProt Consortium (2011). "Ongoing and future developments at the Universal Protein Resource." Nucleic Acids Res 39:D214-D219.21051339
- Scheer, M., Grote, A., Chang, A., Schomburg, I., Munaretto, C., Rother, M., Sohngen, C., Stelzer, M., Thiele, J., Schomburg, D. (2011). "BRENDA, the enzyme information system in 2011." Nucleic Acids Res 39:D670-D676.21062828
- Herrgard, M. J., Swainston, N., Dobson, P., Dunn, W. B., Arga, K. Y., Arvas, M., Bluthgen, N., Borger, S., Costenoble, R., Heinemann, M., Hucka, M., Le Novere, N., Li, P., Liebermeister, W., Mo, M. L., Oliveira, A. P., Petranovic, D., Pettifer, S., Simeonidis, E., Smallbone, K., Spasic, I., Weichart, D., Brent, R., Broomhead, D. S., Westerhoff, H. V., Kirdar, B., Penttila, M., Klipp, E., Palsson, B. O., Sauer, U., Oliver, S. G., Mendes, P., Nielsen, J., Kell, D. B. (2008). "A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology." Nat Biotechnol 26:1155-1160.18846089
- Inoue, Y., Watanabe, K., Shimosaka, M., Saikusa, T., Fukuda, Y., Murata, K., Kimura, A. (1985). "Metabolism of 2-oxoaldehydes in yeasts. Purification and characterization of lactaldehyde dehydrogenase from Saccharomyces cerevisiae." Eur J Biochem 153:243-247.3908097
|
|---|
| Synthesis Reference: | Hsieh, Chun Lung; Houng, Jer Yiing. Preparation of D-lactic acid from D,L-lactic acid ester using wheat germ or pancreatic lipase. U.S. (1997), 5 pp. |
|---|
| External Links: | |
|---|