| Identification |
|---|
| YMDB ID | YMDB00090 |
|---|
| Name | Citicoline |
|---|
| Species | Saccharomyces cerevisiae |
|---|
| Strain | Baker's yeast |
|---|
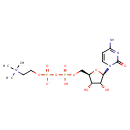
| Description | Citicoline, also known as CDP-colina or nicholin, belongs to the class of organic compounds known as pyrimidine ribonucleoside diphosphates. These are pyrimidine ribonucleotides with diphosphate group linked to the ribose moiety. Citicoline is an extremely weak basic (essentially neutral) compound (based on its pKa). Citicoline exists in all eukaryotes, ranging from yeast to humans. In yeast, citicoline is involved in the metabolic pathway called ether lipid metabolism pathway. |
|---|
| Structure | |
|---|
| Synonyms | - Audes
- CDP-choline
- Cereb
- Choline 5'-cytidine diphosphate
- Choline cytidine diphosphate
- Citicholine
- Citicoline
- Citidoline
- Citifar
- Colite
- Corenalin
- Cyscholin
- Cytidine 5-diphosphate-trihydrogen
- Cytidine 5'-(choline diphosphate)
- Cytidine 5'-(cholinyl pyrophosphate)
- Cytidine 5'-diphosphate choline
- Cytidine 5'-diphosphocholine
- Cytidine choline diphosphate
- Cytidine diphosphate choline
- Cytidine diphosphate choline ester
- Cytidine diphosphocholine
- Cytidine diphosphorylcholine
- cytidine-5' diphosphocholine
- cytidine-5'-pyrophosphate-hydroxycholine
- Cytidoline
- Difosfocin
- Emicholine F
- Ensign
- Haocolin
- Hornbest
- Neucolis
- Nicholin
- Nicolin
- Niticolin
- P-hydroxide[2-(trimethylammonio)ethyl] ester
- Reagin
- Recofnan
- Recognan
- Rexort
- Sintoclar
- Somazina
- Somazine
- Suncholin
- [2-CYTIDYLATE-o'-phosphonyloxyl]-ethyl-trimethyl-ammonium
- CDP-Colina
- Citicolina
- Citicolinum
- Citidin difosfato de colina
- Cyticholine
- Cytidindiphosphocholin
- Cytidine 5'-diphosphoric choline
- [2-CYTIDYLic acid-o'-phosphonyloxyl]-ethyl-trimethyl-ammonium
- Cytidine 5'-(choline diphosphoric acid)
- Cytidine 5'-(cholinyl pyrophosphoric acid)
- Cidifos
- Diphosphate choline, cytidine
- Choline, CDP
- CDP Choline
- 5'-Diphosphocholine, cytidine
- Cytidine 5' diphosphocholine
- Choline, cytidine diphosphate
|
|---|
| CAS number | 987-78-0 |
|---|
| Weight | Average: 488.324
Monoisotopic: 488.107330718 |
|---|
| InChI Key | RZZPDXZPRHQOCG-OJAKKHQRSA-N |
|---|
| InChI | InChI=1S/C14H26N4O11P2/c1-18(2,3)6-7-26-30(22,23)29-31(24,25)27-8-9-11(19)12(20)13(28-9)17-5-4-10(15)16-14(17)21/h4-5,9,11-13,19-20H,6-8H2,1-3H3,(H3-,15,16,21,22,23,24,25)/t9-,11-,12-,13-/m1/s1 |
|---|
| IUPAC Name | {2-[({[(2R,3S,4R,5R)-5-(4-amino-2-oxo-1,2-dihydropyrimidin-1-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl phosphono)oxy]ethyl}trimethylazanium |
|---|
| Traditional IUPAC Name | [2-({[(2R,3S,4R,5R)-5-(4-amino-2-oxopyrimidin-1-yl)-3,4-dihydroxyoxolan-2-yl]methoxy(hydroxy)phosphoryl phosphono}oxy)ethyl]trimethylazanium |
|---|
| Chemical Formula | C14H26N4O11P2 |
|---|
| SMILES | [H]O[C@@]1([H])[C@@]([H])(O[C@]([H])(C([H])([H])OP(=O)(O[H])OP([O-])(=O)OC([H])([H])C([H])([H])[N+](C([H])([H])[H])(C([H])([H])[H])C([H])([H])[H])[C@@]1([H])O[H])N1C([H])=C([H])C(=NC1=O)N([H])[H] |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as pyrimidine ribonucleoside diphosphates. These are pyrimidine ribonucleotides with diphosphate group linked to the ribose moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Nucleosides, nucleotides, and analogues |
|---|
| Class | Pyrimidine nucleotides |
|---|
| Sub Class | Pyrimidine ribonucleotides |
|---|
| Direct Parent | Pyrimidine ribonucleoside diphosphates |
|---|
| Alternative Parents | |
|---|
| Substituents | - Pyrimidine ribonucleoside diphosphate
- Pentose phosphate
- Pentose-5-phosphate
- Glycosyl compound
- N-glycosyl compound
- Phosphocholine
- Monosaccharide phosphate
- Organic pyrophosphate
- Pentose monosaccharide
- Aminopyrimidine
- Pyrimidone
- Monoalkyl phosphate
- Hydropyrimidine
- Monosaccharide
- Organic phosphoric acid derivative
- Phosphoric acid ester
- Pyrimidine
- Imidolactam
- Alkyl phosphate
- Tetrahydrofuran
- Quaternary ammonium salt
- Tetraalkylammonium salt
- Heteroaromatic compound
- Secondary alcohol
- 1,2-diol
- Organoheterocyclic compound
- Oxacycle
- Azacycle
- Organic nitrogen compound
- Organonitrogen compound
- Organopnictogen compound
- Amine
- Alcohol
- Organic oxide
- Organooxygen compound
- Organic oxygen compound
- Organic salt
- Hydrocarbon derivative
- Organic zwitterion
- Primary amine
- Aromatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aromatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Charge | 0 |
|---|
| Melting point | Not Available |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Water Solubility | Not Available | PhysProp | | LogP | Not Available | PhysProp |
|
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations | - endoplasmic reticulum
- cytoplasm
|
|---|
| Organoleptic Properties | Not Available |
|---|
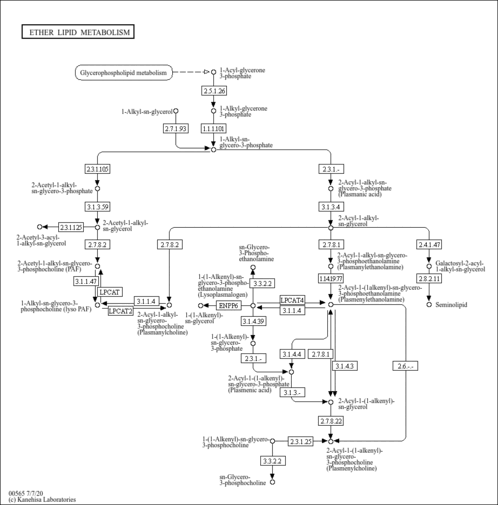
| SMPDB Pathways |
|
|---|
| KEGG Pathways | |
|---|
| SMPDB Reactions |
|
|---|
| KEGG Reactions | |
|---|
| Concentrations |
|---|
| Intracellular Concentrations | Not Available |
|---|
| Extracellular Concentrations | Not Available |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-00fr-7649200000-859d046834f1fbd3d535 | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-00yi-7947012000-9f83d07fb5a59a93e770 | JSpectraViewer | | LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-0006-0001900000-a9f38ff6b185b45f04b3 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-03e9-0590000000-b021677d064d65810ee0 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-001i-0900000000-7570bbb10dd9fa288442 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Positive | splash10-000i-0000900000-8b411373962b073b4d22 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) 30V, Positive | splash10-000i-0001900000-6e7cc437ad79eb13c032 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Positive | splash10-01qi-1932200000-9c3803cefdd9c6285a7c | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Negative | splash10-004i-6505900000-3d534aab6e40915c3642 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Negative | splash10-004i-6505900000-fc13e5ad403e7faf8218 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , negative | splash10-004i-6505900000-3d534aab6e40915c3642 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , negative | splash10-004i-6505900000-fc13e5ad403e7faf8218 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , positive | splash10-000i-0000900000-8b411373962b073b4d22 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , positive | splash10-000i-0001900000-6e7cc437ad79eb13c032 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , positive | splash10-01qi-1932200000-4a7ae92c6e127d2ce453 | JSpectraViewer | MoNA | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-2900100000-0ce672506aae2d1e5eef | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03di-3900000000-c05f3e5b6782e32eeebb | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03di-8900000000-17466c845e643c77969b | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-01p9-0501900000-be567596c9f980cea8c2 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03di-3904100000-4c2e6535887540a12444 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-03fu-9600000000-e42f82159eabb4ec781a | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 2D NMR | [1H,13C] 2D NMR Spectrum | Not Available | JSpectraViewer |
|
|---|
| References |
|---|
| References: | - UniProt Consortium (2011). "Ongoing and future developments at the Universal Protein Resource." Nucleic Acids Res 39:D214-D219.21051339
- Herrgard, M. J., Swainston, N., Dobson, P., Dunn, W. B., Arga, K. Y., Arvas, M., Bluthgen, N., Borger, S., Costenoble, R., Heinemann, M., Hucka, M., Le Novere, N., Li, P., Liebermeister, W., Mo, M. L., Oliveira, A. P., Petranovic, D., Pettifer, S., Simeonidis, E., Smallbone, K., Spasic, I., Weichart, D., Brent, R., Broomhead, D. S., Westerhoff, H. V., Kirdar, B., Penttila, M., Klipp, E., Palsson, B. O., Sauer, U., Oliver, S. G., Mendes, P., Nielsen, J., Kell, D. B. (2008). "A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology." Nat Biotechnol 26:1155-1160.18846089
- Friesen, J. A., Park, Y. S., Kent, C. (2001). "Purification and kinetic characterization of CTP:phosphocholine cytidylyltransferase from Saccharomyces cerevisiae." Protein Expr Purif 21:141-148.11162399
- Percy, A. K., Carson, M. A., Moore, J. F., Waechter, C. J. (1984). "Control of phosphatidylethanolamine metabolism in yeast: diacylglycerol ethanolaminephosphotransferase and diacylglycerol cholinephosphotransferase are separate enzymes." Arch Biochem Biophys 230:69-81.6324684
- McMaster, C. R., Bell, R. M. (1994). "Phosphatidylcholine biosynthesis in Saccharomyces cerevisiae. Regulatory insights from studies employing null and chimeric sn-1,2-diacylglycerol choline- and ethanolaminephosphotransferases." J Biol Chem 269:28010-28016.7961735
- Hjelmstad, R. H., Bell, R. M. (1987). "Mutants of Saccharomyces cerevisiae defective in sn-1,2-diacylglycerol cholinephosphotransferase. Isolation, characterization, and cloning of the CPT1 gene." J Biol Chem 262:3909-3917.3029130
|
|---|
| Synthesis Reference: | Fujio, Tatsuro. New production method of useful substances using ATP regeneration system. Kiraru Tekunoroji no Shintenkai (2001), 187-198. |
|---|
| External Links: | |
|---|